Research Article
Genotyping of the Caprine Kappa-casein Variants in Egyptian Breeds
Department of Cell Biology, National Research Center, Dokki, Egypt
Sahar Ahmed
Department of Cell Biology, National Research Center, Dokki, Egypt
Caprine k-CN was first isolated by Zittle and Custer (1966) and its complete amino acid sequence of 171 residues was established by Mercier et al. (1976). The nucleotide sequence of the cDNA and the promotor region of the gene have been reported (Coll et al., 1993, 1995).
Kappa casein (k-CN) is the milk protein essential for micelle formation and stabilization and influences the manufacturing properties of milk. It differs from other caseins in its solubility over a broad range of calcium ion concentrations and contains a hydrophilic C-terminal region (Mercier et al., 1973).
Cheese making is based on the cleavage of the k-CN Phe105-Met106 peptide bond by enzymes or heat (Gutiérrez et al., 1996). This clearrage isolates the polar domain and leads to micelle coagulation k-CN which is considered to be monomorphic in sheep (Moioli et al., 1998) and widely polymorphic in cattle (Kaminski, 1996).
Due to the linkage between casein genes, it is necessary to take into account k-CN genetic variation for milk protein variants in breeding programs. The bovine k-CN genotype is already listed in some artificial insemination catalogues and selection for k-CN B is practiced in several countries. The milk cows carrying the B allele of k-CN contains a smaller and more homogeneous micelle size, with an elevated concentration of k-CN in milk resulting in higher cheese yield (Ng-Kwai Hang, 1998).
In the present study, forty four animals were analyzed to detect the genetic variants of kappa-casein in four breeds of goat reared in Egypt, Baladi (11 animals), Damascus (12 animals), Sahrawi (11 animals) and Zaraibi (10 animals).
Genomic DNA Extraction
Genomic DNA was extracted from whole blood of 44 goat animals by phenol-chloroform method (John et al., 1991). Ten milliliter of blood taken on EDTA was mixed with 25 mL cold sucrose-triton and the volume was completed to 50 mL by autoclaved double distilled water. The nuclear pellet was suspended in lysis buffer with sodium dodecyl sulfate and proteinase K and incubated overnight in a shaking water-bath at 37°C.
Nucleic acids were extracted once with phenol, saturated with Tris-EDTA (TE) buffer followed by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and this was followed by extraction with chloroform-isoamyl alcohol (24:1). To the final aqueous phase, sodium acetate and cold 95% ethanol were added. The tubes were agitated gently to mix the liquids and a fluffy white ball of DNA was formed.
The DNA was finally dissolved in an appropriate volume of 1X TE buffer. DNA concentrations were determined and diluted to the working concentration of 50 ng μL-1, which is suitable for polymerase chain reaction.
Polymerase Chain Reaction (PCR)
Primes Kb1, Kb2 and 13F have been used for genotyping exon 4 of k-CN in Egyptian goat breeds for detecting four caprine k-CN genetic variants (Yahyaoui et al., 2001, 2003). Kb1 and Kb2 were used to differentiate between A or B and C variants, whereas Kb2 and 13F were used to differentiate between A and E alleles.
| • | Kb1: 5’- TGT GCT GAG TAG GTA TCC TAG TTA TGG -3’ |
| • | Kb2: 5’- GCG TTG TCC TCT TTG ATG TCT CCT TAG -3’ |
| • | 13F: 5’- TCC CAA TGT TGT ACT TTC TTA ACA TC -3’ |
A PCR cocktail consists of 1.0 μM first and second primers and 0.2 mM dNTPs, 10 mM Tris (pH 9), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin (w/v), 0.1% Triton X-100 and 1.25 units of Taq polymerase. The cocktail was aliquot into tubes with 100 ng DNA of goat. The reaction ran in a Perkin Elmar apparatus. The reaction was cycled for 1 min at 95°C, 1.30 min at 63°C and 1.30 min at 72°C for 35 cycles.
RFLP and Agarose Gel Electrophoresis
Two different restriction enzymes were used in this study, Alw44I was used to detect A or B and C alleles, whereas HaeIII was used to detect A and E alleles. Twenty microliter of PCR product were digested with 10 units of each restriction enzyme used in this study in a final reaction volume 25 μL. The reaction mixture was incubated at 37°C in water bath over night. After restriction digestion, the restricted fragments were analyzed by electrophoresis on 2.5% agarose/1X TBE gel stained with ethidium bromide. The 100 bp ladder was used as molecular size marker. The bands were visualized under UV light and photographed with yellow filter on black and white film.
The casein genes are organized as a cluster including αS1, αS2, β-CN and k-CN locus (Ferretti et al., 1990; Threadgill and Womack, 1990). Kappa casein gene comprises five exons which more than 90% of the coding region for mature protein is contained in the penultimate exon (exon 4) (Yahyaoui et al., 2001). The analyzed region of exon 4 contains the major part of the coding sequence for mature protein (141 amino acids over a total 171) (Yahyaoui et al., 2003). Primes I3F, Kb1 and Kb2 have been used for genotyping exon 4 in Egyptian goat breeds.
The PCR of the 459 bp region of k-CN exon 4 was amplified by kb1 and kb2 primers for detecting A or B and C alleles. The digested fragments by Alw44I produced two restriction patterns at 381 and 78 bp for allele C, while the PCR products from A and B alleles remained intact (Fig. 1).
The PCR of the 645 bp region of k-CN exon 4 was amplified by Kb2 and I3F primers for detecting A and E alleles. The digested fragments with HaeIII produced two fragments at 416 and 229 bp for allele A, while allele E displayed by restriction patterns at 366, 229 and 50 bp. Allele B was detected by combining the results of digestion with Alw44I and HaeIII endonucleases.
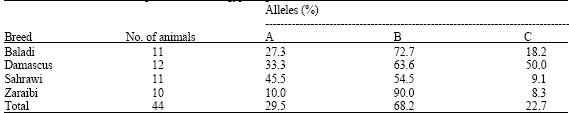
The result of k-CN genetic variants in Egyptian goat showed that the appearance of allele A in 13 of 44 tested animals with a frequency 29.5%, allele B was displayed with a high frequency 68.2% (30 animals) and allele C was appeared in 10 animals (22.7%). The appearance of each allele was in heterozygous or homozygous genotype. The E allele was not present in all 44 tested animals.
Among different breeds, we found that the allele B is the most common allele in the four tested breeds and appeared with the maximum frequency in Zaraibi breed (90%) whereas the allele C was appeared at low frequencies in Sahrawi (9.1%) and Zaraiby (8.3%) breeds. Allele A was displayed in different frequencies ranged from 45.5% (Sahrawi) to 10% (Zaraibi) (Table 1).
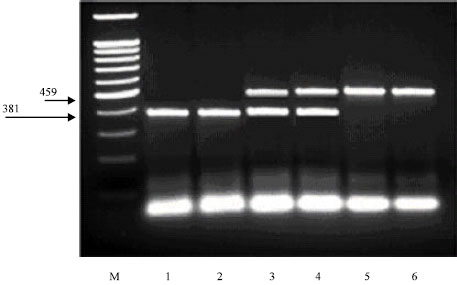 | |
| Fig. 1: | DNA electrophoretic pattern obtained after digestion of PCR amplified goat kappa-casein products with Alw44I, M: 100 bp ladder marker, Lanes 1 and 2 showed the digested fragment of 381 bp (specific for C allele). Lans 3 and 4 showed the undigested fragment of 459 bp (specific for A or B allele) and the digested fragment of 381 bp (specific for C allele). Lanes 5 and 6 showed the undigested fragment of 459 bp (specific for A or B allele) |
| Table 1: | A, B and C allele frequencies in four Egyptian goat breeds |
 | |
This could be explained due to the caprine k-CN B allele is the ancestral allele, while k-CN C allele is more divergent and the A variant shows an intermediate similarity. The difference between k-CN A and k-CN B involves only one amino acid substitution at position 119, where the valine in variants A is substituted by isoleucine in variant B. The k-CN A differs from the C variant in the following amino acid substitution: valine for isoleucine at position 65 and 119, alanine for valine at position 156 and serine for proline at position 159. The first amino acid substitution (position 65) accurs in the N-terminal region (caseinomacropeptide) (Yahyaoui et al., 2001).
The Italian goat breeds, Garganica, Jonica and Maltese recorded no evidence for C and E alleles (Chessa et al., 2003). The C allele is present at low frequencey (1%) in the Spanish Murciano-Granadina and Canaria breeds and not found in the Malaguena or the Payoya breeds. The frequency of this allele is present with 11% in the French Saaen breed (Yahyaoui et al., 2001). The frequencies of A and B alleles reported high percentages compared to other alleles in different Europian breeds (Caroli et al., 2001; Yahyaoui et al., 2001; Angolillo et al., 2002; Chessa et al., 2003; Yahyaoui et al., 2003; Jann et al., 2004).
Identification of the genotypes of Egyptian goat milk caseins αS1 (Ahmed, 2006), αS2 (Othman and Ahmed, 2006) and kappa casein (k CN) in this study, will enable to establish well strategy for breeding the Egyptian goat contributed in filling the nutritious gap in milk and milk productions. Especially, goat is considered as an animal with low price, highly prolificacy (produce twins or triples) and has the ability to the adaptation in different environment compared to buffalo or cattle.
This study is conducted using DNAs provided by the project titled “Genetic diversity of indigenous sheep and goat breeds in Egypt”, funded by the Egyptian Academy of Research and Technology.