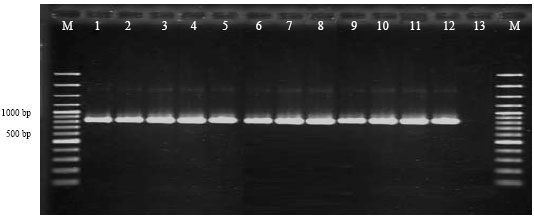
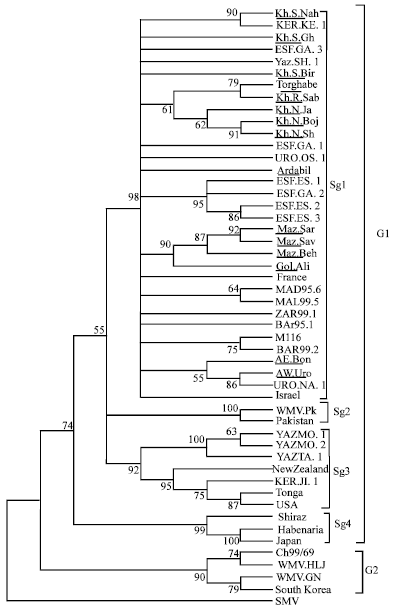
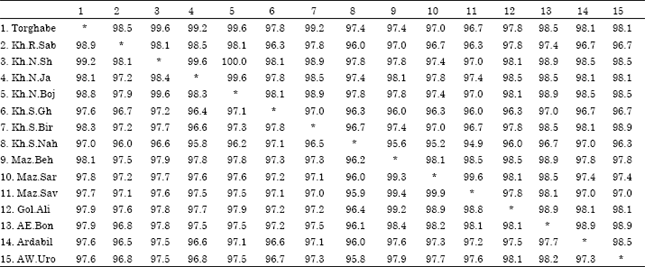
Research Article
Diagnosis and Molecular Variability of Watermelon mosaic virus Isolates from North, East, North-east and North-west Regions of Iran
Department of Plant Pathology, College of Agriculture, Ferdowsi University of Mashhad, Khorasan, Iran