Research Article
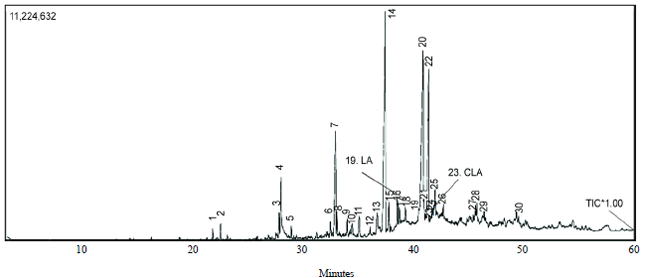
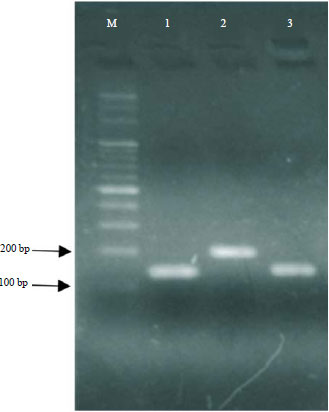
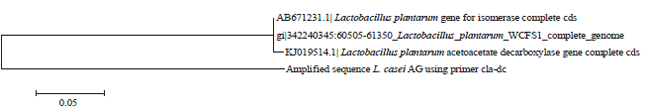
Conjugated Linoleic Acid Synthesis in Milk Fermented with Lactobacillus casei Strain AG
Faculty of Animal Science, Universitas Gadjah Mada, Yogyakarta, Indonesia
Ari Surya Sukarno
Graduate Studies on Biotechnology, Universitas Gadjah Mada, Yogyakarta, Indonesia
Kafaah Estancia
Faculty of Animal Science, Universitas Gadjah Mada, Yogyakarta, Indonesia
Donny Widianto
Faculty of Agriculture, Universitas Gadjah Mada, Yogyakarta, Indonesia
Indratiningsih
Faculty of Animal Science, Universitas Gadjah Mada, Yogyakarta, Indonesia