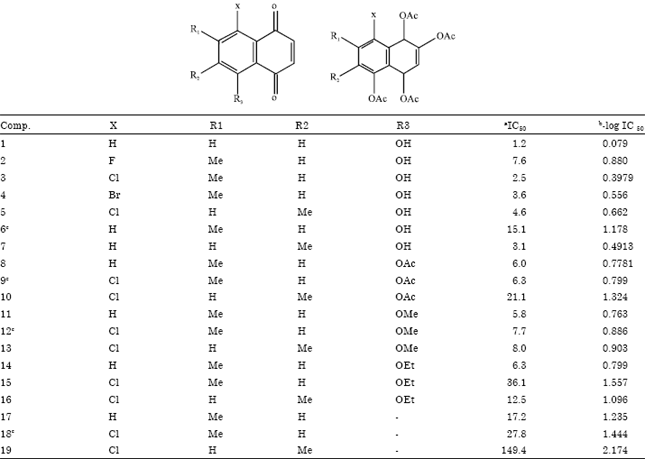
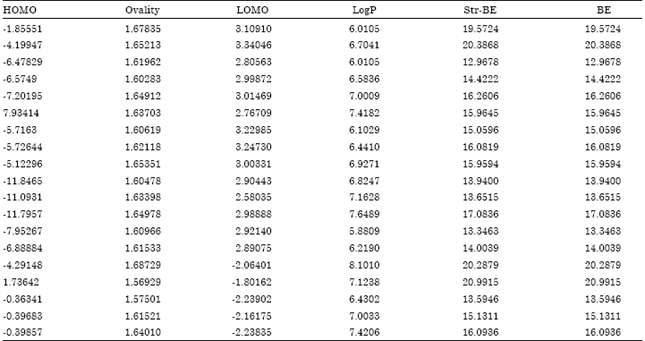
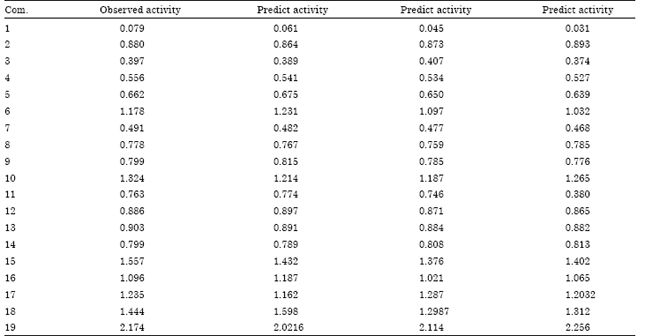
Research Article
2D Qsar Study of 7-Methyljuglone Derivatives: An Approach to Design Anti Tubercular Agents
School of Pharmacy, Devi Ahilya Vishwavidyalaya, Indore (M.P) 452001, India
Smita Sharma
Department of Chemistry, Chodhary Dilip Singh Kanya Mahavidyalya Bhind (M.P)-477001, India