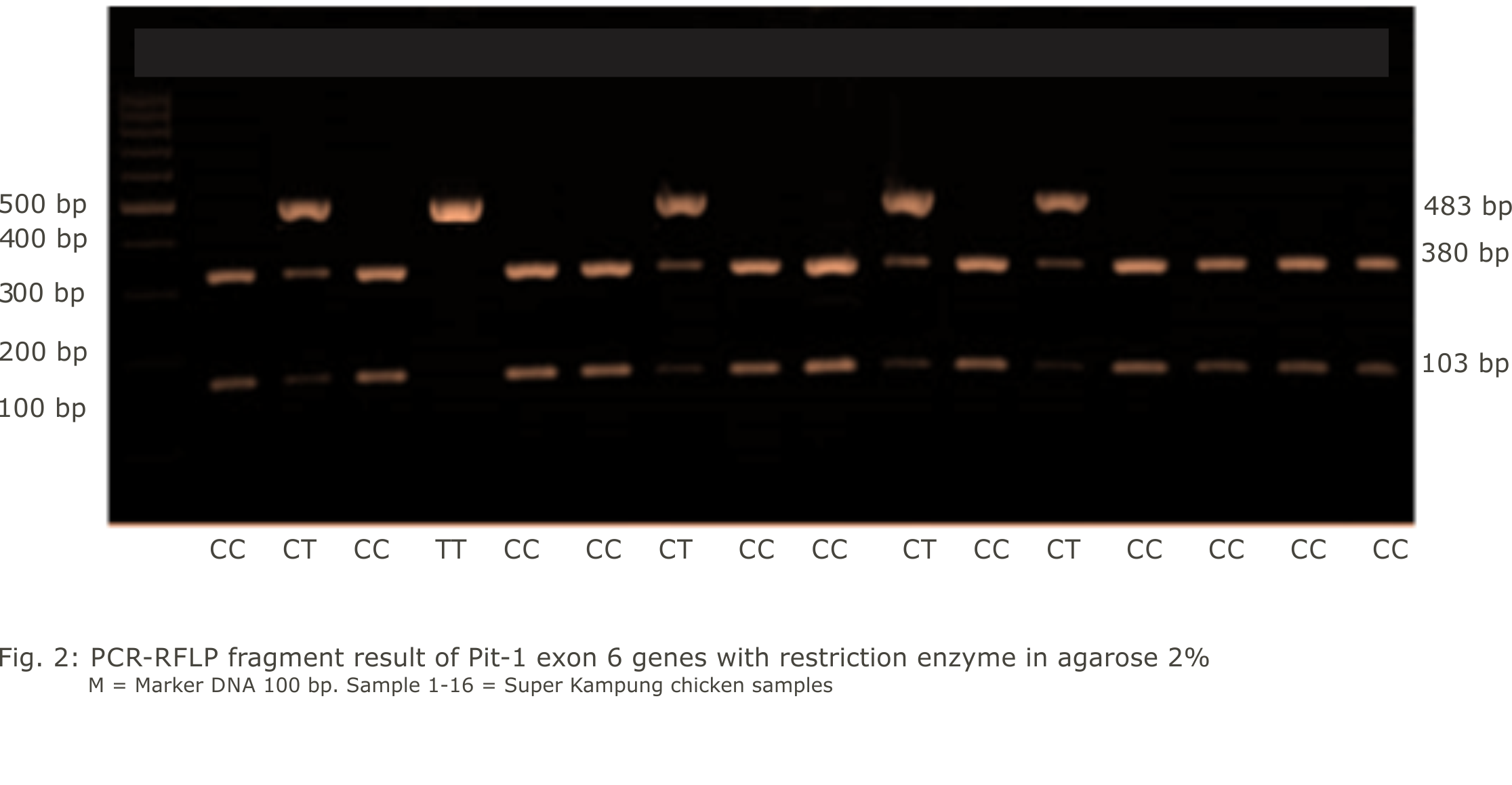
Research Article
Pituitary-Specific Transcription Factor Gene Polymorphism and its Associations with Body Weight of Super Kampung Chickens Aged Thirteen Weeks
Faculty of Animal Science, University of Mataram, Jalan Majapahit No. 62 Mataram, Lombok 83125, Indonesia
LiveDNA: 62.18907
B. Indarsih
Faculty of Animal Science, University of Mataram, Jalan Majapahit No. 62 Mataram, Lombok 83125, Indonesia
N.K.D. Haryani
Faculty of Animal Science, University of Mataram, Jalan Majapahit No. 62 Mataram, Lombok 83125, Indonesia
M. Muhsinin
Faculty of Animal Science, University of Mataram, Jalan Majapahit No. 62 Mataram, Lombok 83125, Indonesia