Research Article
Microbiological Qualities of Goat Milk Obtained under Different Milking Systems at a Smallholder Dairy Farm in Yogyakarta, Indonesia
Faculty of Animal Science, Universitas Gadjah Mada, Jl Fauna No. 3, Bulaksumur, Yogyakarta 55281, Indonesia
Yustina Yuni Suranindyah
Faculty of Animal Science, Universitas Gadjah Mada, Jl Fauna No. 3, Bulaksumur, Yogyakarta 55281, Indonesia
Widodo
Faculty of Animal Science, Universitas Gadjah Mada, Jl Fauna No. 3, Bulaksumur, Yogyakarta 55281, Indonesia
LiveDNA: 62.23055









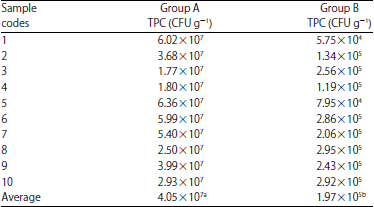
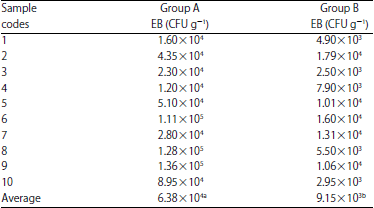
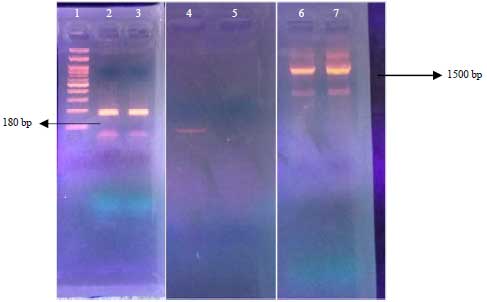
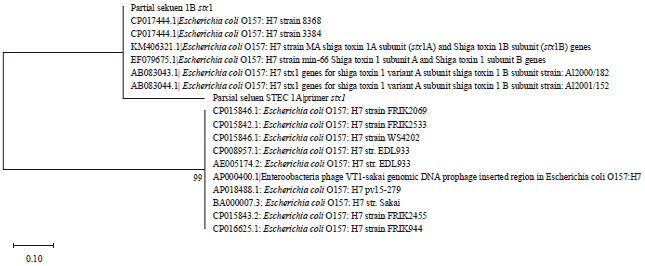
Daniel Kahsu Reply
thank you for help me