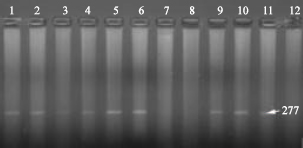
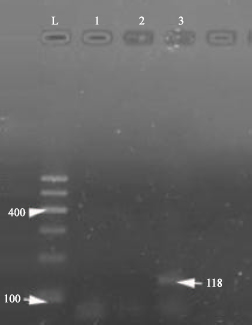
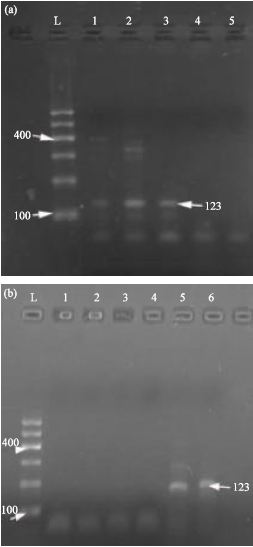
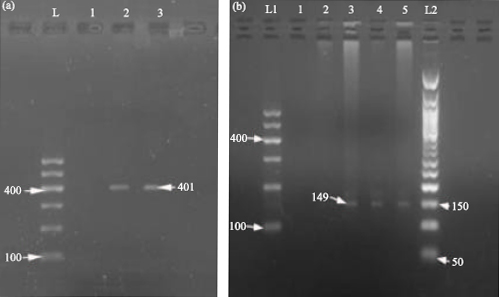
Research Article
Detection of Genetically Modified Maize and Soybean Food Products in the Jordanian Market
Princess Sumaya University for Technology, Amman, Jordan
H. Al-Rousan
Royal Scientific Society, Amman, Jordan
B.O. Hayek
Princess Sumaya University for Technology, Amman, Jordan
M.A. Ibrahim
Princess Sumaya University for Technology, Amman, Jordan