Research Article
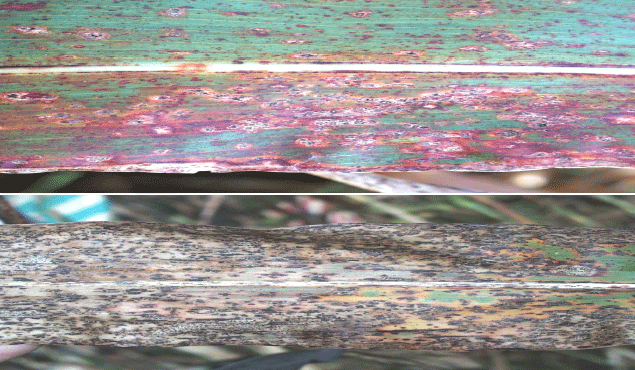
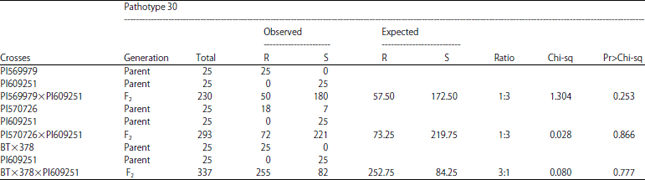
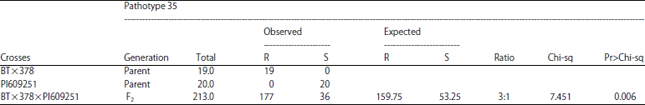
Inheritance of Resistance of Three Sorghum Lines to Pathotypes of Colletotrichum sublineola, Causal Agent of Anthracnose
USDA-ARS, Plains Area Agricultural Research Center, College Station, 77845, Texas
LiveDNA: 1.3546
Hugo Cuevas
USDA-ARS, Tropical Agriculture Research Station, 2200 Pedro Albizu Campos Avenue, Mayaguez, 00680, Puerto Rico
Ramasamy Perumal
Kansas State University, Agricultural Research Center, Hays, 67601, Kansas
LiveDNA: 91.1156
Thomas Isakeit
Department of Plant Pathology and Microbiology, Texas A and M University, College Station, 77843, Texas
Clint Magill
Department of Plant Pathology and Microbiology, Texas A and M University, College Station, 77843, Texas