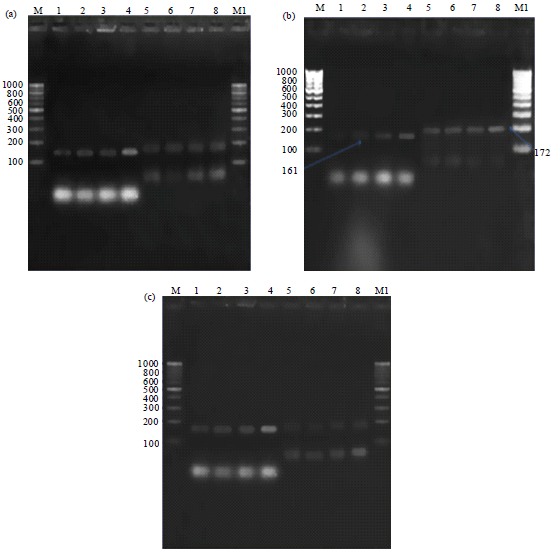
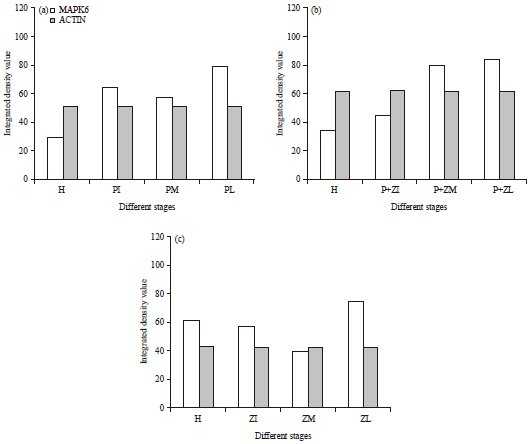
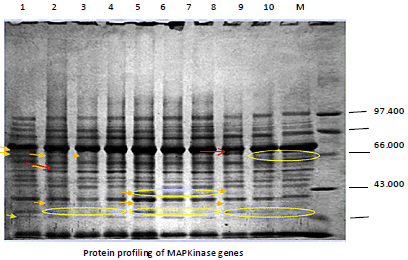
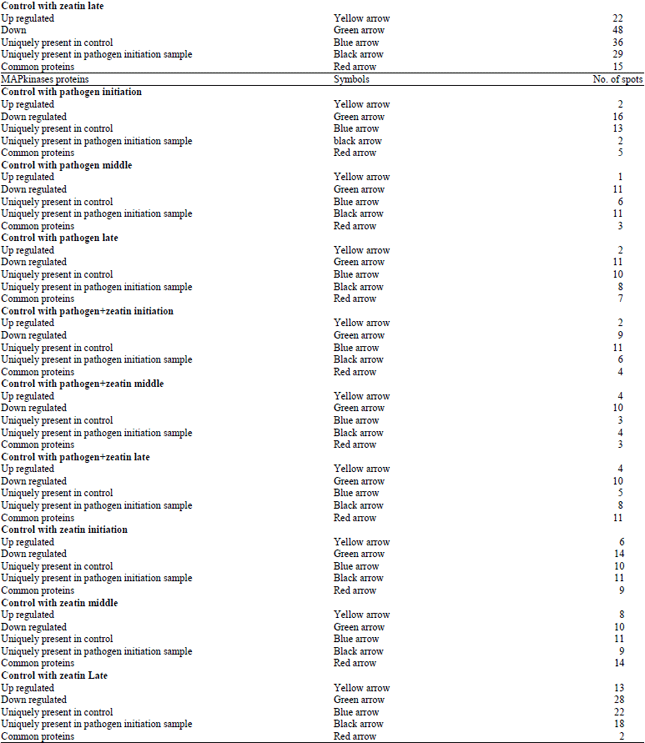
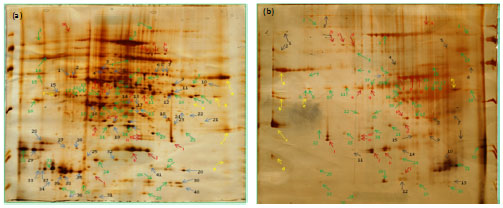
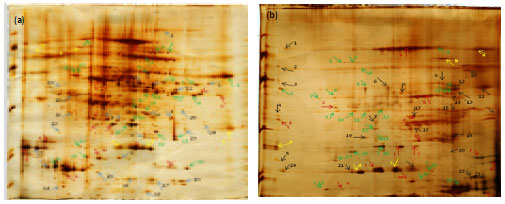
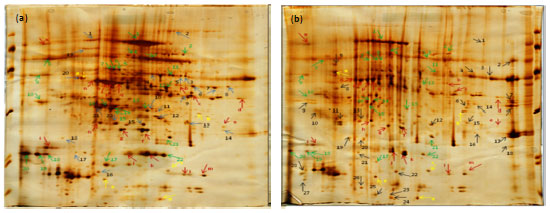
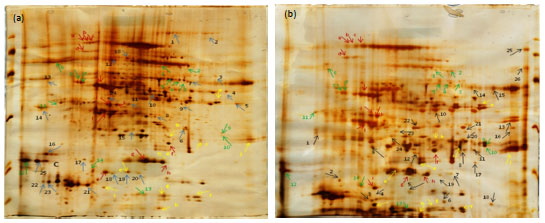
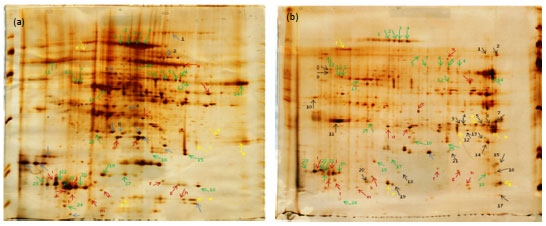
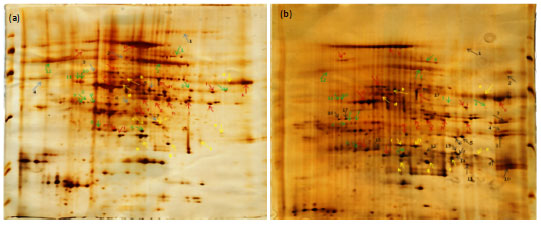
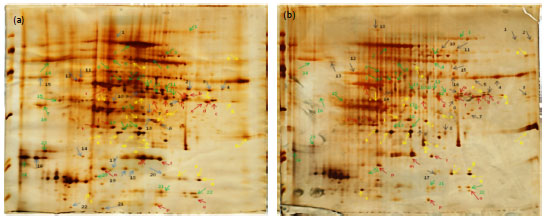
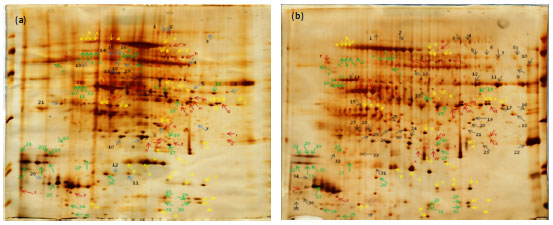
Research Article
ATMPK4 and ATMPK6 Transcript and Protein Profiling in Arabidopsis thaliana Plants Challenged with Zeatin and Alternaria brassicae
Department of Molecular Biology and Genetic Engineering, College of Basic Sciences and Humanities, G.B. Pant University of Agriculture and Technology, Pantnagar, Uttarakhand, 263145, India
M. Baunthiyal
Department of Bioscience and Biotechnology, Banasthali University, PO Banasthali Vidyapith, Rajasthan 304 022, India
D. Pandey
Department of Molecular Biology and Genetic Engineering, College of Basic Sciences and Humanities, G.B. Pant University of Agriculture and Technology, Pantnagar, Uttarakhand, 263145, India
A.K. Gupta
Department of Molecular Biology and Genetic Engineering, College of Basic Sciences and Humanities, G.B. Pant University of Agriculture and Technology, Pantnagar, Uttarakhand, 263145, India