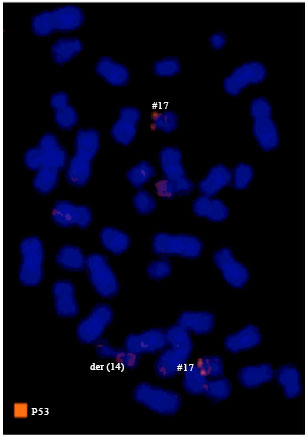
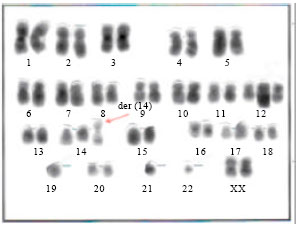
Research Article
Detection P53 Gene Deletion in Hematological Malignancies Using Fluorescence In situ Hybridization: A Pilot Study
College of Medicine, Kufa University, P.O. Box 18, Najaf, Iraq
Alice K. Melconian
College of Medicine, Kufa University, P.O. Box 18, Najaf, Iraq
Ban A. Abdul-Majeed
College of Medicine, Kufa University, P.O. Box 18, Najaf, Iraq
Ali M. Jawad
College of Medicine, Kufa University, P.O. Box 18, Najaf, Iraq