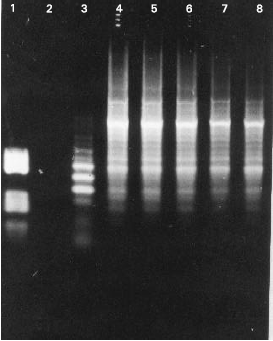
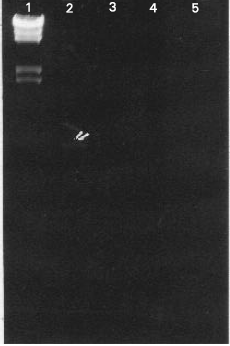
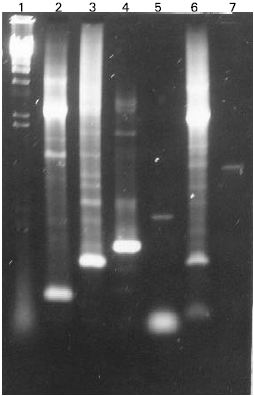
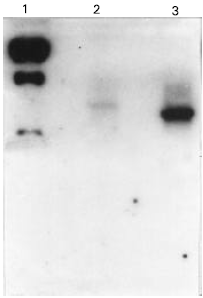
Research Article
PCR Amplification and Cloning of T-DNA Homologous Sequences from Nicotiana glauca
Department of Plant Breeding and Genetics, University of Azad Jammu and Kashmir, Faculty of Agriculture, Rawalakot Azad Jammu and Kashmir, Pakistan
Mukhtar Alam
Pakistan Oil Development Board, Ministry of Food Agriculture and Livestock Islamabad, Pakistan
R.J. K. Narayan
Institute of Biological Science, University of Wales, Aberystwyth, UK
M. Ilyas Khan
Department of Plant Breeding and Genetics, University of Azad Jammu and Kashmir, Faculty of Agriculture, Rawalakot Azad Jammu and Kashmir, Pakistan