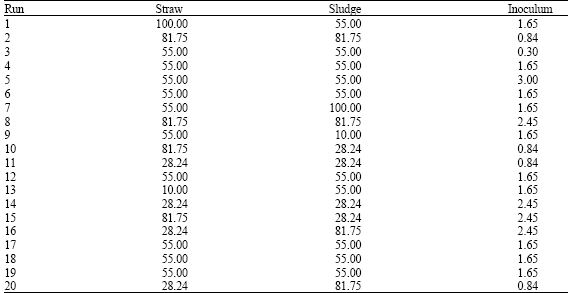
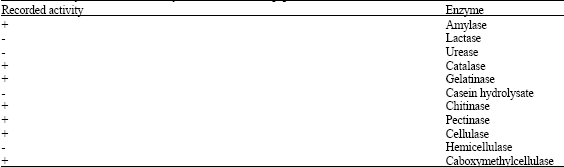
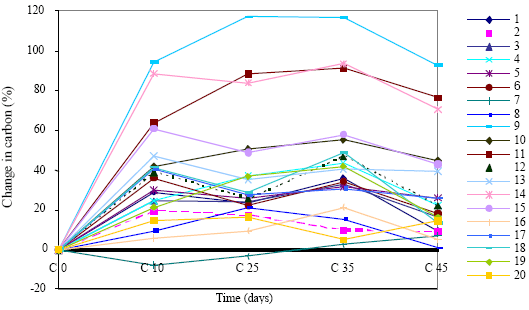
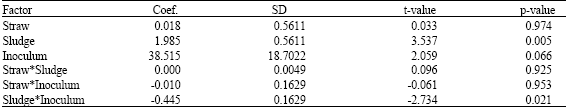
Research Article
Factorial Design for Optimization of Rice Straw Incorporation into Soil Using Micromonospora chalcea
Department of Botany, Faculty of Sciences, Suez Canal University, Ismailia, Egypt
Hesham Abdulla
Department of Botany, Faculty of Sciences, Suez Canal University, Ismailia, Egypt
Ahmed Dewedar
Department of Botany, Faculty of Sciences, Suez Canal University, Ismailia, Egypt