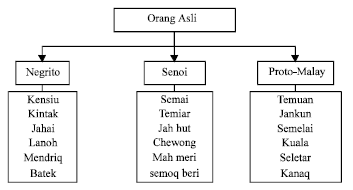
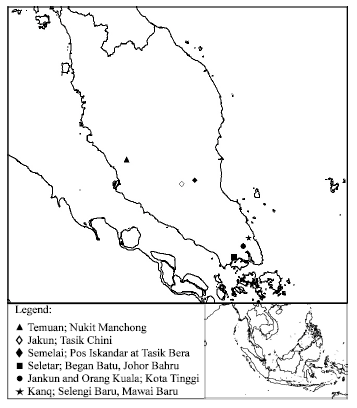
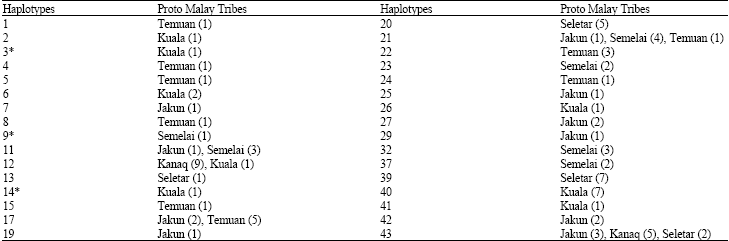
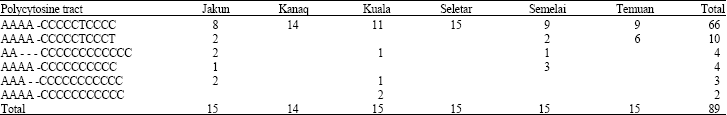
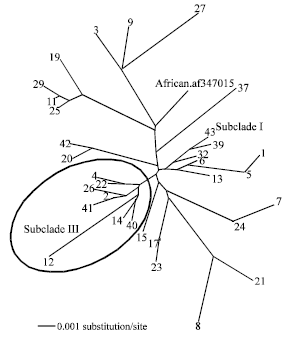
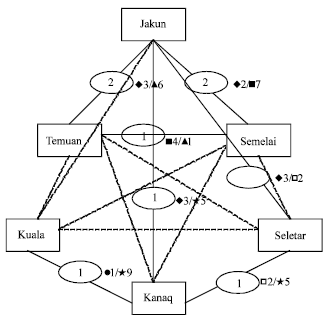
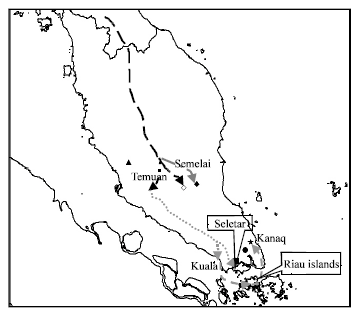
Research Article
Mitochondrial DNA Polymorphism and Phylogenetic Relationships of Proto Malays in Peninsular Malaysia
School of Environmental and Natural Resources Sciences, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Selangor, Malaysia
K.C. Ang
School of Environmental and Natural Resources Sciences, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Selangor, Malaysia
M.C. Mahani
School of Environmental and Natural Resources Sciences, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Selangor, Malaysia
A.W. Shahrom
Forensic Unit, Hospital Universiti Kebangsaan Malaysia, Jalan Ya`acob Latif, Bandar Tun Razak, 56000 Cheras, Kuala Lumpur, Malaysia
B.M. Md-Zain
School of Environmental and Natural Resources Sciences, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Selangor, Malaysia