Research Article
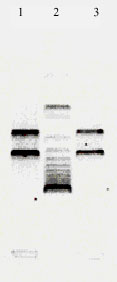
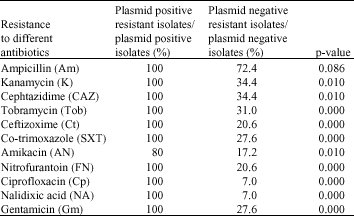
Plasmid Profiling of Klebsiella sp. and its Relation with Antibiotic Resistance at two Hospitals of Urmia (Iran)
Department of Microbiology, Immunology and Genetics, Faculty of Medicine, Urmia University of Medical Sciences, Urmia, Iran
Mir Davood Omrani
Department of Microbiology, Immunology and Genetics, Faculty of Medicine, Urmia University of Medical Sciences, Urmia, Iran
Zahra Sabahi
Department of Microbiology, Immunology and Genetics, Faculty of Medicine, Urmia University of Medical Sciences, Urmia, Iran
Masumeh Mosavi
Department of Microbiology, Immunology and Genetics, Faculty of Medicine, Urmia University of Medical Sciences, Urmia, Iran
Minoo Zartoshti
Department of Microbiology, Immunology and Genetics, Faculty of Medicine, Urmia University of Medical Sciences, Urmia, Iran