Research Article
Evaluation of Some Faba Bean Genotypes against Chocolate Spot Disease Using cDNA Fragments of Chitinase Gene and Some Traditional Methods
Department of Agronomy, Faculty of Agriculture, Cairo University, Giza, Egypt
Noha F. El-Badawy
Plant Pathology Research Institute, ARC, Giza, Egypt
M.M. Mazen
Plant Pathology Research Institute, ARC, Giza, Egypt
H. Abd El-Menem
Plant Pathology Research Institute, ARC, Giza, Egypt









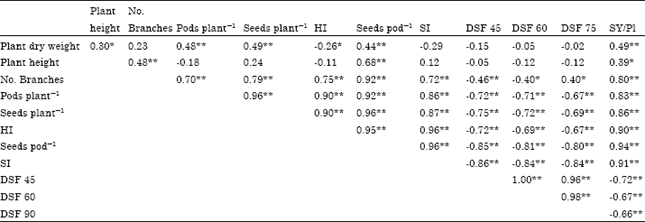
hamdy mohamed abo shedida Reply
This paper is useful l need all paper related this subject
Dr. Samir Abo Hegazy
Dear Dr. Hamdy
I am so sorry because i can't send it now for you, because i am not in Egypt now, may be after 2 monthes when i come back to Egypt, send you these papers
hamdy mohamed gamal eldeen aboshedida
dear dr samir/iam hamdy i requested from you the papers ralated this subject before and you tell me you will send them to me when you come back to egypt and i should be grateful to you if you send me these things