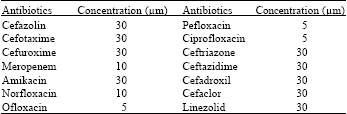
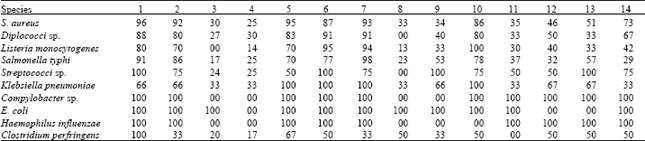
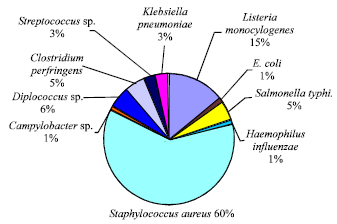
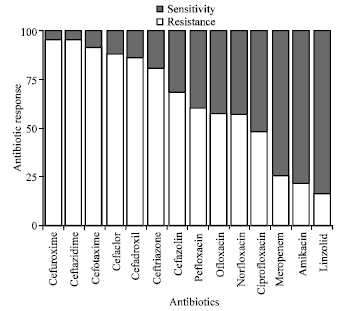
Research Article
Prevalence, Profile and Antibiotic Susceptibility Pattern of Bacterial Isolates from Blood
Department of Microbiology, S.G.B. Amravati University, Amravati-444602, India
D.V. Dhanorkar
Department of Microbiology, S.G.B. Amravati University, Amravati-444602, India
S.R. Gulhane
Department of Microbiology, S.G.B. Amravati University, Amravati-444602, India
M.N. Dudhane
Department of Biochemistry, Dr. P.D.M. Medical College, Amravati-444603, India