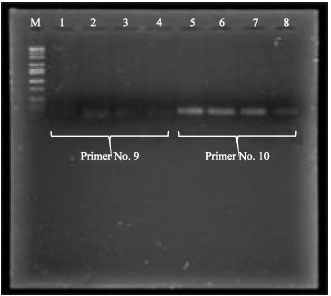
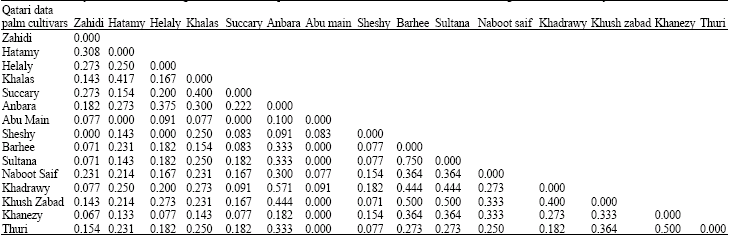
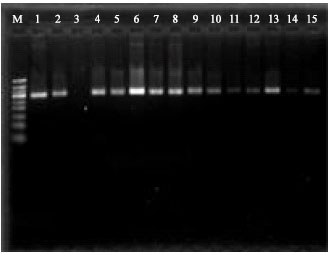
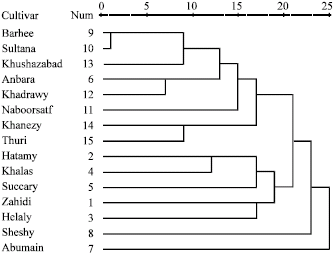
Research Article
Molecular Phylogeny Of Qatari Date Palm Genotypes Using Simple Sequence Repeats Markers
Department of Biological and Environmental Sciences, College of Arts and Sciences, Qatar University, Doha, Qatar
Asmaa Y. Al-Qaradawi
Department of Biological and Environmental Sciences, College of Arts and Sciences, Qatar University, Doha, Qatar