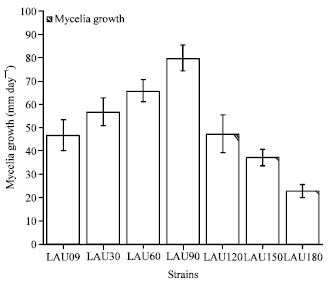
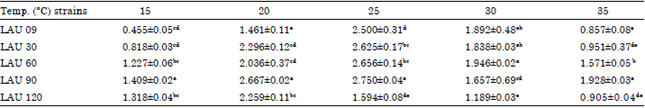
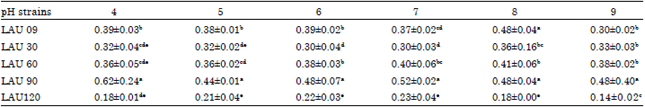
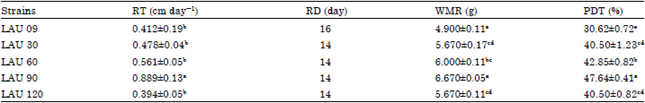
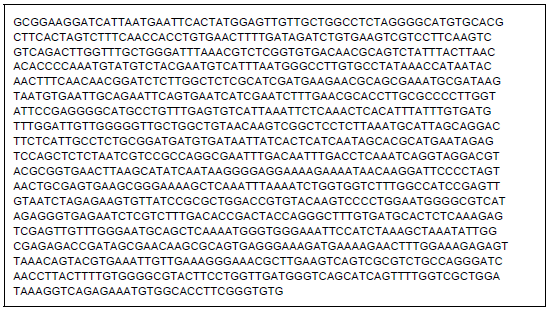Research Article
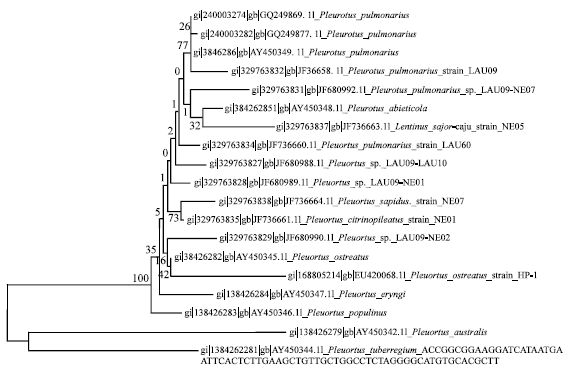
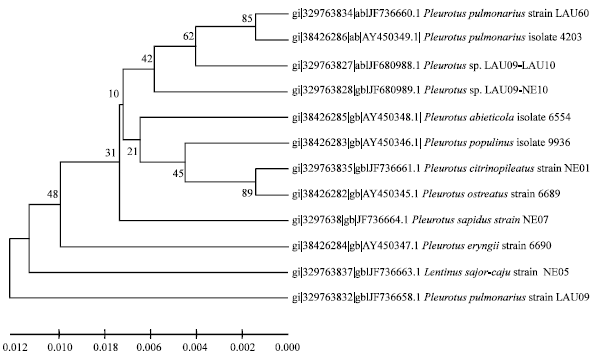
Improvement of Pleurotus pulmonarius Lau 09 Through Mutation for Yield Performance
Department of Pure and Applied Biology, Ladoke Akintola University of Technology, P.M.B. 4000, Ogbomoso, Nigeria
J.K. Oloke
Department of Pure and Applied Biology, Ladoke Akintola University of Technology, P.M.B. 4000, Ogbomoso, Nigeria
A.K. Bordoloi
North East Institute of Science and Technology, CSIR, Jorhat-785006, Assam, India
M. Barooah
Assam Agricultural University Jorhat-13, Assam, India
T.C. Bora
North East Institute of Science and Technology, CSIR, Jorhat-785006, Assam, India