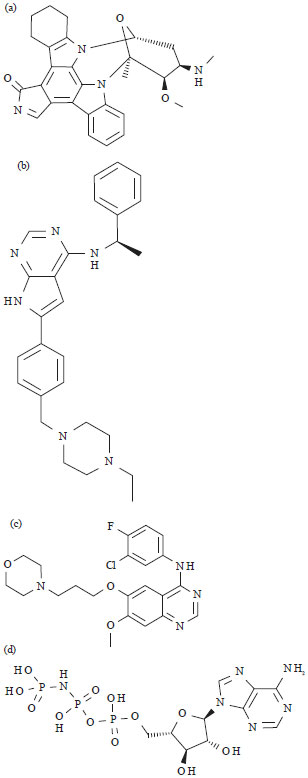
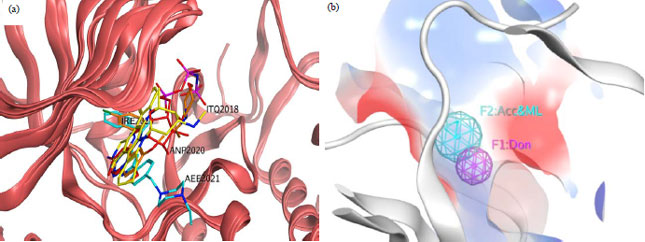
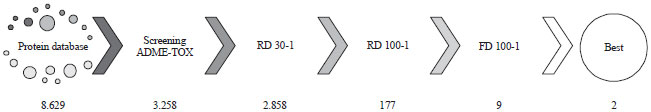
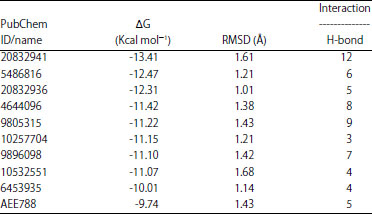
Research Article
In silico Identification of Peptide as Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Lung Cancer Treatment
Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Indonesia, Depok, Indonesia
Eka G. Ningsih
Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Indonesia, Depok, Indonesia
Usman S.F. Tambunan
Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Indonesia, Depok, Indonesia
LiveDNA: 62.13001