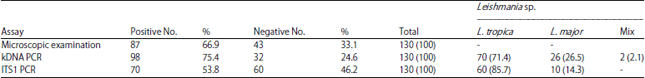Research Article
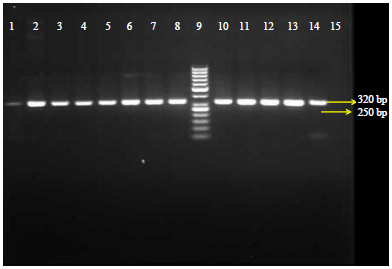
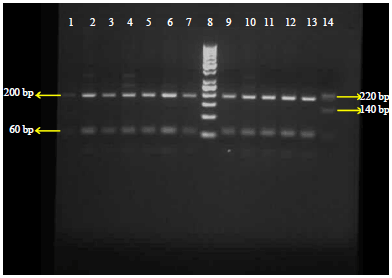
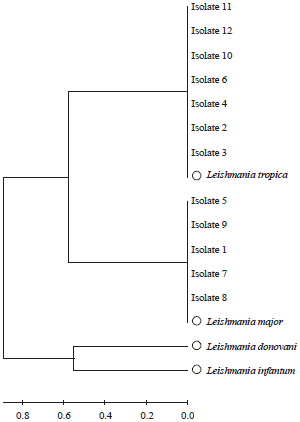
Molecular Diagnosis of Clinical Isolates of Cutaneous Leishmaniasis Using ITS1 and KDNA Genes and Genetic Polymorphism of Leishmania in Kashan, Iran
Department of Parasitology and Mycology, School of Medicine, Kashan University of Medical Sciences, Kashan, Iran
Sima Rasti
Department of Parasitology and Mycology, School of Medicine, Kashan University of Medical Sciences, Kashan, Iran
Mahdi Delavari
Department of Parasitology and Mycology, School of Medicine, Kashan University of Medical Sciences, Kashan, Iran
LiveDNA: 98.13174
Abbas Doroodgar
Department of Parasitology and Mycology, School of Medicine, Kashan University of Medical Sciences, Kashan, Iran