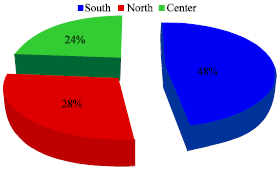
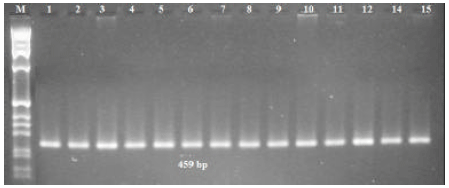
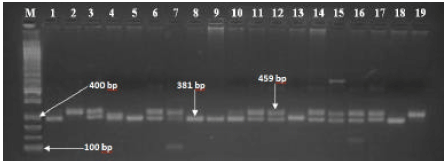
Research Article
Kappa Casein Gene Polymorphism in Local Tunisian Goats
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia
A. Ben Gara
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia
H. Selmi
Regional Field crops Research Center Beja, Tunisia
Z. Ammari
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia
C. Bouheni
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia
M. Ben Larbi
National Agricultural Institute of Tunisia, Tunisia
M. Hammami
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia
M. Amraoui
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia
M. Kamoun
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia
H. Rouissi
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia
B. Rekik
Department of Animal Production, School of Higher Education in Agriculture of Mateur, Tunisia