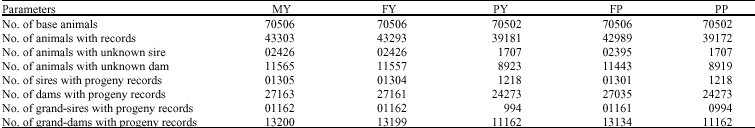
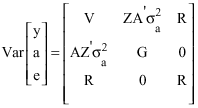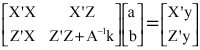Research Article
Prediction Breeding Value and Genetic Parameter in Iranian Holstein Bulls for Milk Production Traits
Department of Animal Science, College of Agriculture, IslamicAzad University, Science and Research Branch, Tehran, Iran
Naser Emam Jomeh Kashan
Department of Animal Science, College of Agriculture, IslamicAzad University, Science and Research Branch, Tehran, Iran
Mohammad Reza Nassiry
Department of Animal Science, Ferdowsi University of Mashhad, Iran
Rasol Vaez Torshizi
Department of Animal Science, Tarbiat Modarres University, Tehran, Iran