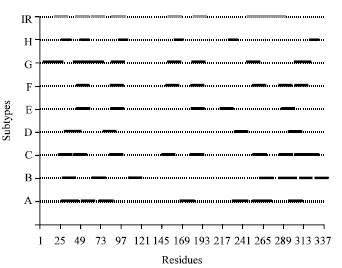
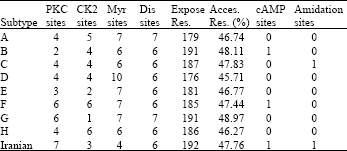
Research Article
Computational Comparison of T-cell Epitopes of gp120 of Iranian HIV-1 with Different Subtypes of the Virus
Department of Biology, College of Sciences, Shiraz University, Shiraz 71454, Iran
Reza Mohammadzadegan
Department of Biology, College of Sciences, Shiraz University, Shiraz 71454, Iran