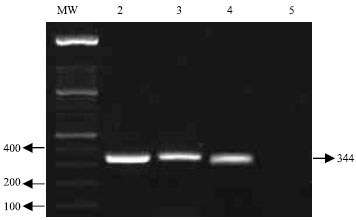
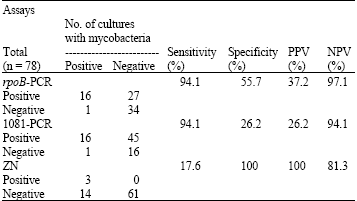
Research Article
Evaluation of Different Primer Sets for the Rapid Diagnosis of Tuberculosis
Department of Microbiology, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran
P. Maleknejad
Department of Microbiology, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran
A. Bahador
Department of Microbiology, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran
H. Peeri- Dogaheh
Faculty of Medicine, Ardebil University of Medical Sciences, Ardebil, Iran
M. Y. Alikhani
Faculty of Medicine, Hamedan University of Medical Sciences, Hamedan, Iran
R. Radmanesh-Ahsani
Department of Microbiology, Faculty of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran