Research Article
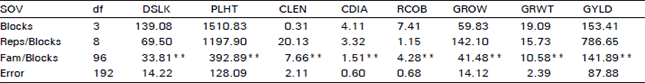
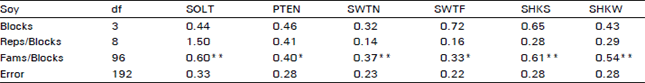
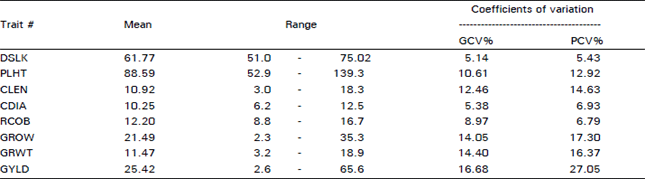
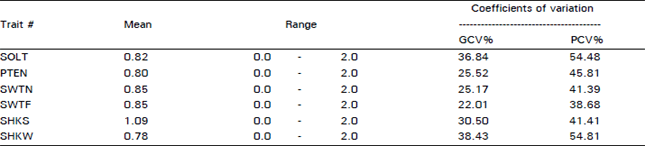
Variability For Grain Yield, its Components and Quality Traits in a Sweet Corn Population
Department of Plant Breeding and Genetics, University of Agriculture, Faisalabad, Pakistan
Syed Sadaqat Mehdi
Department of Plant Breeding and Genetics, University of Agriculture, Faisalabad, Pakistan