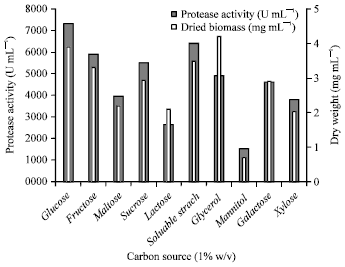
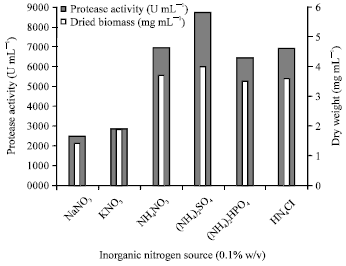
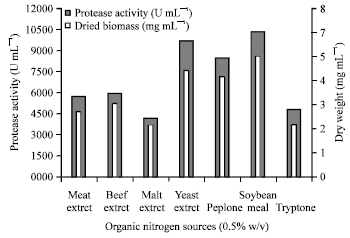
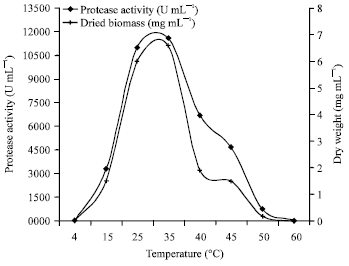
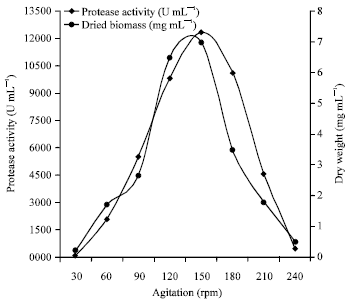
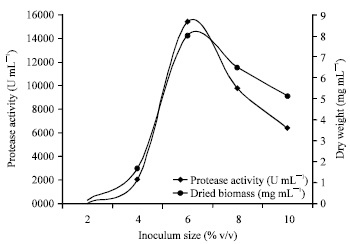
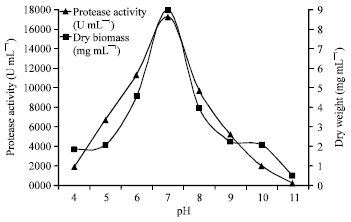
Research Article
Characterization of Marine Streptomyces carpaticus and Optimization of Conditions for Production of Extracellular Protease
Department of Biotechnology, College of Science and Technology, Andhra University, Visakhapatnam 530003, India
K. SivaKumar
Department of Biotechnology, College of Science and Technology, Andhra University, Visakhapatnam 530003, India
A. Swathi
Department of Biotechnology, College of Science and Technology, Andhra University, Visakhapatnam 530003, India
Y.S.Y.V. Jagan Mohan
Department of Biotechnology, College of Science and Technology, Andhra University, Visakhapatnam 530003, India
T. Ramana
Department of Biotechnology, College of Science and Technology, Andhra University, Visakhapatnam 530003, India