Research Article
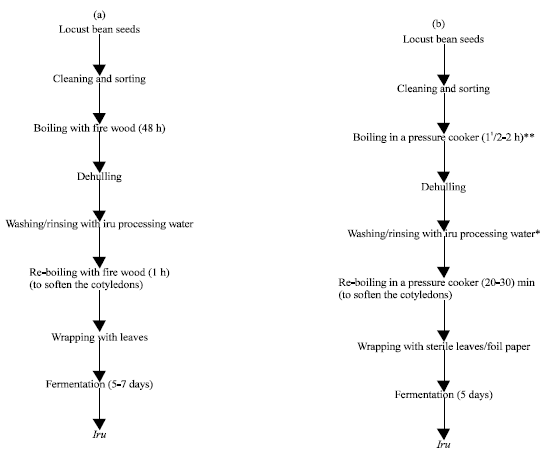
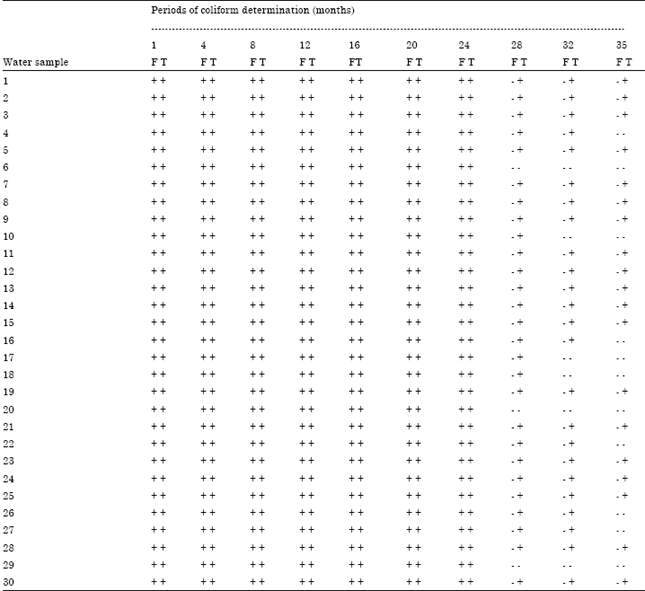
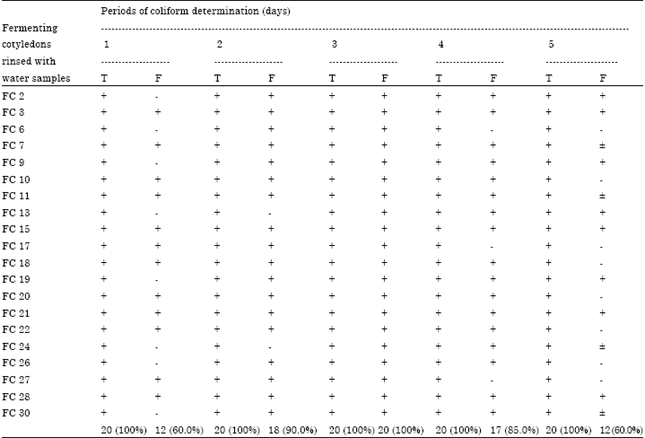
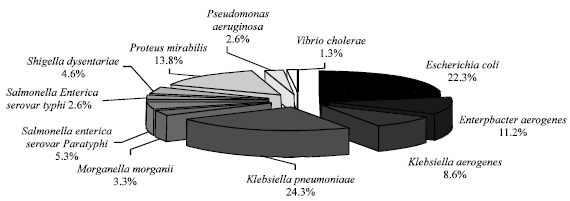
Processing-Water as Source of Gram-negative Foodborne Indicator Bacteria in Traditionally-produced Iru
Applied Microbiology and Infectious Diseases, Department of Microbiology, Faculty of Science, University of Ibadan, Nigeria
J.N. Okereh
Applied Microbiology and Infectious Diseases, Department of Microbiology, Faculty of Science, University of Ibadan, Nigeria