Research Article
Parametric Estimation of the Immunes Proportion based on BCH Model and Exponential Distribution using Left Censored Data
Institute for Mathematical Research, Universiti Putra Malaysia, Malaysia
Mohd Rizam A. Bakar
Department of Mathematics, Institute for Mathematical Research, Universiti Putra Malaysia, Malaysia
Noor Akma Ibrahim
Department of Mathematics, Institute for Mathematical Research, Universiti Putra Malaysia, Malaysia
Habshah Midi
Department of Mathematics, Institute for Mathematical Research, Universiti Putra Malaysia, Malaysia









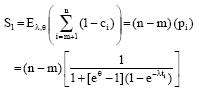
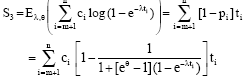
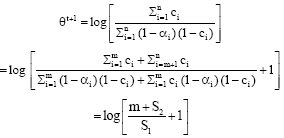
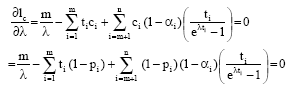
Raed Falah Reply
Very interesting analysis and the implementation of the EM algorithm is so nice, but no real data or simulation has involved. We are interesting to see how we can apply the proposed procedure on some real data.