Research Article
Rapid Automatic Segmentation and Visualization of Teeth in CT-Scan Data
Control and Intelligent Processing Center of Excellence, Faculty of Engineering, School of Electrical and Computer Engineering, University of Tehran, Tehran, Iran
R.A. Zoroofi
Control and Intelligent Processing Center of Excellence, Faculty of Engineering, School of Electrical and Computer Engineering, University of Tehran, Tehran, Iran
G. Shirani
Departmentof Oral and Maxillofacial Surgery, Faculty of Dentistry Medical Science, University of Tehran, Tehran, Iran









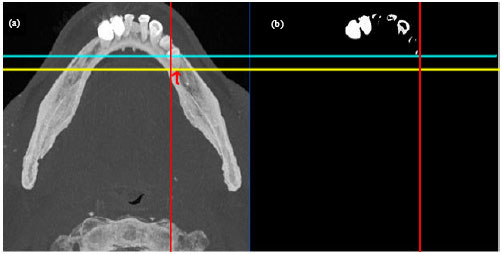
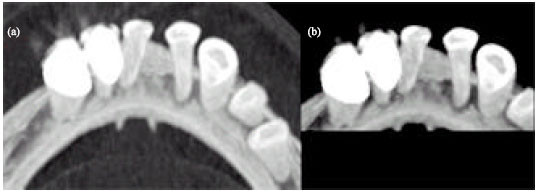
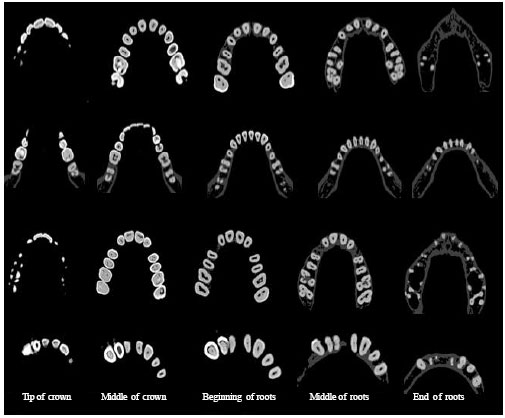
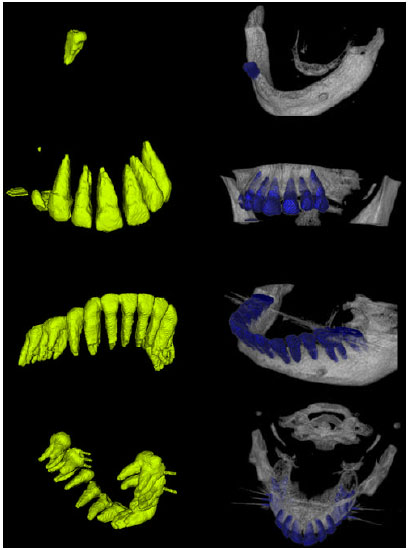
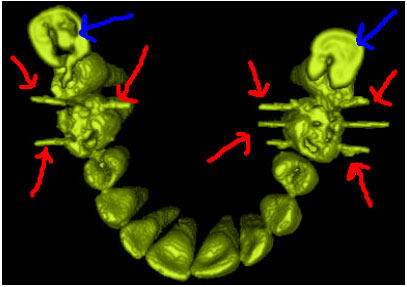
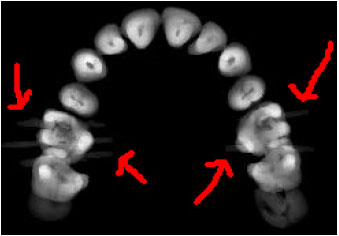
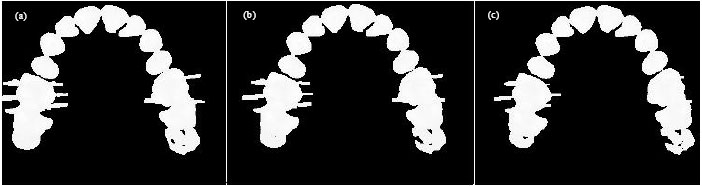

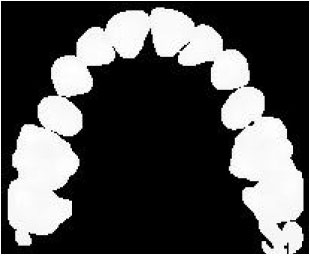
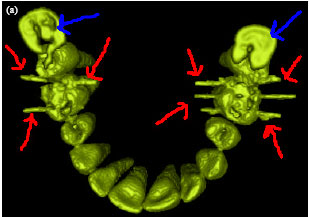
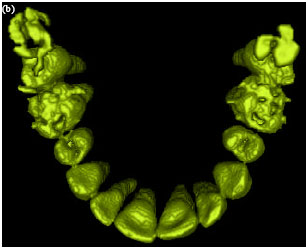
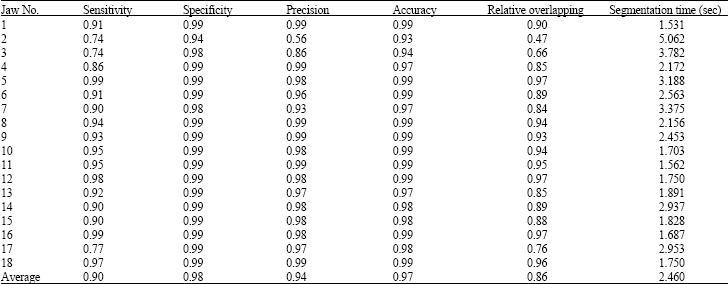
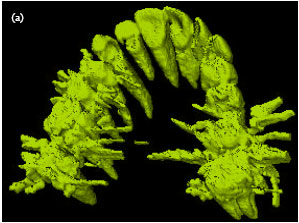
Sharma Mayank Reply
Hello Sir,
I am doing my masters from IIT Madras and on same project.
I had gone through your paper on same topic and liked it, liked the approach.
But to actually implement it on matlab, can't figure it out how to approach.
Can you send me the codes for this segmentation and reconstruction, this will be great help.
H. Akhoondali
Hello,
The algorithm has been implemented using C++ not Matlab. Unfortunately, it is not possible to give the codes.
Kind regards