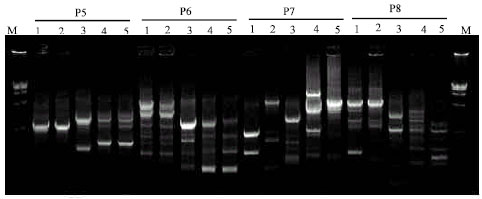
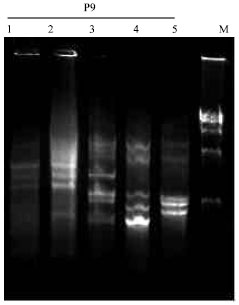
Research Article
Differentiation of Bacillus thuringiensis and Escherichia coli by the Randomly Amplified Polymorphic DNA Analysis
Department of Nucleic Acid Research, Genetic Engineering and Biotechnology Research Institute, Mubarak City for Scientific Research and Technology Applications, New Borg El-Arab City 21934, Alexandria, Egypt
Tian-Hua Huang
Department of Cell Biology and Medical Genetics, Shantou University, Medical College, 22 XinLing Road, Shantou, Guangdong 515031, Peoples Republic of China
Bahy Ahmed Ali
Department of Cell Biology and Medical Genetics, Shantou University, Medical College, 22 XinLing Road, Shantou, Guangdong 515031, Peoples Republic of China
Qing- Dong Xie
Department of Cell Biology and Medical Genetics, Shantou University, Medical College, 22 XinLing Road, Shantou, Guangdong 515031, Peoples Republic of China