Research Article
Molecular Characterization of Tomato Yellow Leaf Curl Virus (TYLCV) Infecting Pepper and Common Bean
Department of Biotechnology, Collage of Agriculture and Food Science, King Faisal University, 31982 Al-Ahsa, Saudi Arabia
Tomato Yellow Leaf Curl Virus (TYLCV) causes one of the most devastating viral diseases in tomato plants (Solanum lycopersicum) worldwide (Czosnek and Laterrot 1997; Hanssen et al., 2010). The TYLCV (family Geminiviridea, genus Begomovirus) was first observed in tomato plants more than 70 years ago in the eastern Mediterranean (Antignus and Cohen, 1994; Lapidot and Friedman, 2002; Polston et al., 2006). Recently, seven genera have been recognized within the family Geminiviridea according to insect vector, genome arrangement and sequences identity (Brown et al., 2012; Adams et al., 2013; Martinez-Ayala et al., 2014). The genus Begomovirus is considered the most prevalent worldwide within this family. These viruses infect dicotyledonous plants and are transmitted in nature by the whitefly Bemisia tabaci (De Barro et al., 2011). The majority of the members of this genus are clustered in two main phylogenetic clades called old and new world (Briddon et al., 2010). Most of the old world members have monopartite genome with one genomic component of ~2.8 kb (DNA-A). while the new world members have bipartite genomes with two genomic components of ~2.6 kb each (designated DNA-A and DNA-B), except Tomato leaf deformation virus (ToLDeV) recently reported from Peru as monopartite begomovirus (Sanchez-Campos et al., 2013; Melgarejo et al., 2013; Martinez-Ayala et al., 2014). Sub-viral molecules have been found associated with some monopartite viruses from the Eastern Hemisphere called betasatellites (DNA-β), which encode a single protein on the complementary-sense strand, referred to as βC1 (Briddon et al., 2002, 2003; Idris et al., 2012). The βC1 protein has been shown to function as a virulence factor of disease severity and symptom development (Briddon et al., 2001; Cui et al., 2004).
Recently, TYLCV has been detected in tomato plants from different locations in Saudi Arabia (Alhudiab et al., 2014). Recent studies based on molecular analyses has resulted in the characterization and elucidation of the genomic sequence (Alhudiab et al., 2014). The TYLCV was observed to infect not only tomato but also other cultivated crops including (Capsicum spp.) (Morilla et al., 2005; Polston et al., 2006) and common bean (Phaseolus vulgaris) (Navas-Castillo et al., 1999; Lapidot and Friedman, 2002; Papayiannis et al., 2007; Segundo et al., 2008; Kamaal et al., 2015). In addition, TYLCV infects various weeds such as henbit (Lamium amplexicaule) (Kil et al., 2014). There is little information and evidence available on TYLCV infection of pepper and common bean crops. Most of this evidence reported for TYLCV infection of C. annuum and P. vulgaris while only one study reported C. frutescens to be a host for TYLCV (Polston et al., 2006). In this study, this study report the molecular characterization of 2 different strains of TYLCV that naturally infect C. frutescens and P. vulgaris in Saudi Arabia. This study provides definitive evidence based on molecular data and nucleotide sequence that beta satellite (DNA-β) was associated with TYLCV isolated from P. vulgaris.
Sample collection: Plant samples of pepper (Capsicum frutescens) and common bean (Phaseolus vulgaris) showing yellowing and leaf curling symptoms were collecting from two different locations of Saudi Arabia, Jizan (Southwest of Saudi Arabia) and Hail (North of Saudi Arabia) during 2012-2013. Samples were kept in cool boxes after collecting from the field and stored at -80°C or used directly for DNA extraction.
Total DNA extraction and PCR detection: Total DNA was extracted from the leaves samples using DNaesy plant mini extraction kit (Qiagen) and according to its manufacturer’s guidelines. Polymerase Chain Reaction (PCR) was used to detect Begomovirus infection in the collected samples using the degenerate primer pair AVcore/ACcore (Brown et al., 2001). These primers amplify fragments of DNA-A of ~580 bp (Brown et al., 2001). The PCR reactions were done in a Techne TC-4000 Thermal Cycler. Each PCR reaction contained 2.5 μL of extracted DNA, 0.25 mM MgCl2, 0.25 mM dNTPs, 2.5 μM of each primer (AVcore/ACcore), 1×enzyme buffer and 0.5 U of Taq DNA polymerase in 25 μL final volume. The PCR amplification program started with an initial denaturing step at 94°C for 2 min, followed by 30 cycles of 94°C for 1 min, 50°C for 2 min and 72°C for 2 min and a final extension step at 72°C for 10 min. The amplified products were determined on 1.5% agarose gels in TBE buffer (tris-boric acid- EDTA) and stained with ethidium bromide (0.5 mg mL–1).
Rolling Circle Amplification (RCA): Total nucleic acids were extracted from symptomatic leaves of pepper and common bean using DNaesy plant mini extraction kit (Qiagen) as mentioned above. Extracted DNA was subjected to TempliPhi amplification (Amersham Biosciences, Piscataway, NJ, USA) and according to the manufacturer’s instructions. In brief, 2 μL (10-20 ng) of total Extracted DNA were dissolved in 5 μL of sample buffer, denatured for 3 min at 95°C and cooled down on ice for 1 min followed by addition of 5 μL reaction buffer and 0.2 μL enzyme mix. Amplification was performed for overnight (16-20 h) at 30°C and the reaction was stopped by heating for 10 min at 65°C to inactivate the enzyme. The amplified DNA was used as a template for the PCR to amplify the full-length genome of tomato yellow leaf curl virus and to detect the occurrence of β satellite.
Full-length amplification of TYLCV-DNA-A: The DNA-A of TYLCV were amplified by PCR using the primer pairs (TYLCV1F 5-ACAGCCCATACAGCAGCCGTGC-3 and TYLCV1R 5-GCTGTCGAAGTTCAGCCTTCGGC-3). The PCR reaction contained 1 μL of RCA product, 0.25 mM MgCl2, 2.5 μM of each primer (TYLCV1F/ TYLCV1R), 0.25 mM dNTPs, 1×enzyme buffer and 0.5 U of HotStar Taq Plus DNA polymerase (Qiagen) in a total volume of 25 μL. The viral DNAs were amplified using amplification program as following; one cycle of denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 2 min and extension at 72°C for 2 min followed by final extension step at 72°C for 10 min. Amplified DNA fragments were electrophoresed in 0.7% agarose gel in TBE buffer and visualized using UV transilluminator after staining with ethidium bromide.
Amplification of DNA-β: To study the presence of DNA-β molecules in the TYLCV infected samples, two consensus outwardly extending primers were used (beta F1, 5-GTTGTCTTGT TTATTATGTTTGTTGGC-3 as forward primer; beta 02, 5-GGTACCTACCCTCCCAGGGGTACAC-3 (Briddon et al., 2002) as reverse primer. The PCR reaction contained 1 μL of RCA product of template DNA, 15 mM MgCl2 buffer, 10 pmol dNTPs, 40 pmol of each primer and 2.5 U Taq polymerase. The reactions were denatured at 94°C for 3 min, followed by 30 cycles at 94°C for 1 min, 55°C for 1 min and 72°C for 1 min, terminating with 10 min at 72°C.
Cloning and sequence analysis: The amplified fragments for DNA-A and DNA-β were ligated into pGEM-T easy cloning vector (Promega, WI, USA) and cloned according to the manufacturer’s instructions. After transformation into the DH5α strain of Escherichia coli, white colonies were screened for the gene of interest by PCR and restriction digestion with EcoRI. General primers Sp6 and T7 were used to sequence the clones of DNA-A and DNA-β fragments in both directions (Macrogen Inc., Seoul, Korea). Alignment analysis were performed using the online BLAST service of the National Center for Biotechnology Information (URL:http//www.ncbi.nlm.nih.gov/BLAST/). The GC content for the two isolates in this sudy was calculated using the Geecee software (http:// weblab.cbi.pku.edu.cn/program.inputForm.do?Program=geecee) and SnapGene viewer software version 3.1.
Phylogenetic and homology analysis: Nucleotide sequences reported in this study were deposited in GenBank under the accession numbers KF435136 (isolate from C. frutescens) and KF444467 (isolate from P. vulgaris). Sequences were retrieved and BLAST analyzed (National Center for Biotechnology Information (URL:http//www.ncbi.nlm.nih.gov/BLAST/). Phylogenetic trees were constructed with the DNAMAN software version 8 (Lynnon, Canada) usingthe neighbor-joining method (Saitou and Nei, 1987) and the Jukes-Cantor distance correction method (Jukes and Cantor, 1969).
Sampling and PCR amplification: Virus infected plants of common bean and pepper collected from the field showed yellowing and leaf curling symptoms (Fig. 1). The plants in early stage showed stunting and the production decreased. The relationship of a begomoviruses with the symptomatic P. vulgaris and C. frutescens was confirmed by PCR using AVcore/ACcore primers as degenerate primers for begomoviruses. The symptomatic P. vulgaris and C. frutescens samples gave bands of the expected size (~580 bp) (Fig. 2a). The RCA was performed to amplify all circular DNA molecules from the extracted DNA for the samples that gave amplicons with AVcore/ACcore primers. As in Fig. 2b the expected size (~2.8 kb) of the full length DNA-A genome was amplified from RCA P. vulgaris and C. frutescens DNA templates. In addition, a fragment (722 bp) of the DNA-β satellite genome was successfully amplified using RCA of P. vulgaris as template (Fig. 2c). This study never found any DNA-β satellite by RCA using C. frutescens as template.
Full genome sequences analysis: Full-length genome sequences of TYLCV-Pep from C. frutescens and TYLCV-GB from P. vulgaris were analyzed and compared with other known sequences in the DNA database that obtained from GenBank (Table 1). The DNA-B was not found by PCR in the two TYLCV isolates using different sets of specific primers. However, DNA-β satellite was found associated with TYLCV-GB when DNA-β specific primers were used. The size of the full length genomes of TYLCV-Pep and TYLCV-GB were 2764 and 2779 bp respectively (Fig. 3).
 | |
| Fig. 1(a-b): | (a) Phaseolus vulgaris and (b) Capsicum frutescens showing yellowing and leaf curling symptoms of Begomovirus infections |
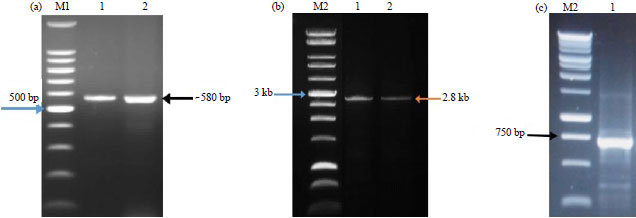 | |
| Fig. 2(a-c): | Agarose gel electrophoresis of PCR amplified products of, (a) Partial coat protein gene, (b) Full-length genome of TYLCV and (c) Partial fragment of DNA-β from the RCA templates of TYLCV-GB and TYLCV-Pep isolates. Lanes M1 = 100 bp marker, M2 = 1 kb ladder, 1 = Sample from common bean (TYLCV-GB) and 2 = Sample from pepper (TYLCV-Pep) |
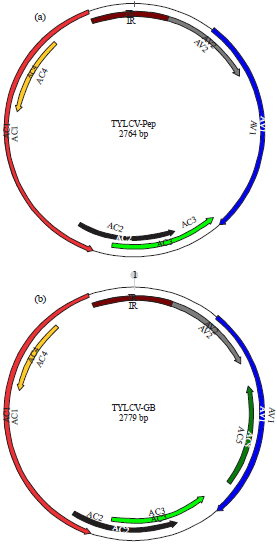
| Table 1: | GenBank accession numbers for tomato yellow leaf curl virus sequences used for sequence analysis |
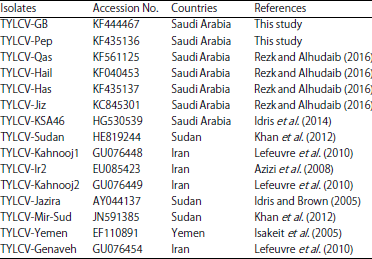 | |
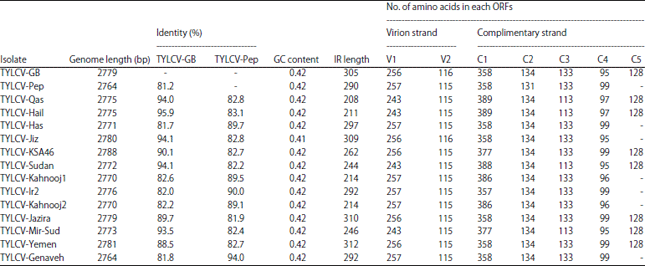
The length of the IR was 290 bp for TYLCV-Pep and 305 bp for TYLCV-GB. The homology and phylogenetic analysis are shown in Fig. 4 and Table 2. The nucleotide identity between TYLCV-GB and TYLCV-Pep was 81.2%. The highest nt identity of TYLCV-GB was 96.9% with TYLCV-Hail, while the highest nt identity of TYLCV-Pep was 94% with TYLCV-Genaveh (isolate from Iran). As in Table 2, six Open Reading Frames (ORFs) were identified in the TYLCV-Pep genome (two in the viral strand and four in viral complementary strand) whereas seven ORFs were determined at the TYLCV-GB genome (two in the viral strand and five in the viral complementary strand). The number of amino acids translated from each ORF in the two isolates is shown in Table 2.
 | |
| Fig. 3(a-b): | Genome size and ORF organization of, (a) TYLCV-Pep and (b) TYLCV-GB using SnapGene viewer 3.1 software |
The GC content was 42% in both TYLCV-GB and TYLCV-Pep; the GC content in all known begomoviruses ranges between 41-42% (Table 2). The IR identity value between TYLCV-GB and TYLCV-Pep was very low (74.6%) whereas it was 98.6% between TYLCV-GB and TYLCV-Hail.
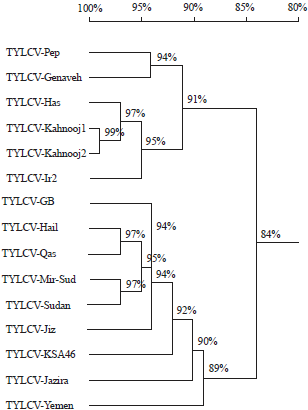 | |
| Fig. 4: | Homology tree showing the identity percentage among the isolated Tomato Yellow Leaf Curl Virus (TYLCV-GB and TYLCV-Pep) and other strains and isolates selected from GenBank and were aligned with the optimal alignment method of DNAMAN |
| Table 2: | Genomic characterization of TYLCV-GB and TYLCV-Pep and other isolate and strains that available in GenBank database |
 | |
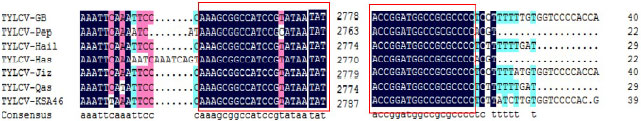 | |
| Fig. 5: | Conserved region of stem-loop structure in the all Saudi TYLCV recorded in GenBank database |
Also this value was 94.5% between TYLCV-Pep and TYLCV-Genaveh. All the Saudi TYLCV isolates contain an identical nucleotide sequence of 42 nucleotides (AAAGCGGCCATCCGT-ATAATATTACCGGATGGCCGCGCCCC) inside the IR region (Fig. 5). This conserved region forms a characteristic stem-loop structure as in all begomoviruses.
Nucleotide sequence analysis of DNA-β: Two pairs of primers beta F1 and beta 02 were used to PCR-amplify 722 nucleotides of DNA β satellite from RCA products as described above. The PCR amplicons were cloned into pGEM-Teasy vector (Promiga) and multiple clones were obtained. One of them was selected and sequenced in both orientations. Alignments and phylogenetic analyses were performed using DNAMAN (version 8) and BLAST). The DNA-β satellite was found associated with TYLCV infected common bean plants but not with TYLCV infected pepper plants. The obtained sequence of DNA-β satellite shows genome arrangement typical of DNA-β satellites associated with TYLCV isolates from Yemen (Idris et al., 2012), with identity not up to 90%. One ORF was identified in the complementary sense known as βC1, predicted to a protein of encode 117 amino acids.
There are a few reports on the incidence of TYLCV infecting pepper and beans. The reports showing susceptibility of Capsicum spp. to TYLCV are conflicting. The inoculation studies in the greenhouse with isolates from Jordan and Israel indicated that C. annuum was not a host for TYLCV (Mansour and Al-Musa, 1992; Polston et al., 2006), in contrast, other a report from Spain indicated that C. annuum is indea a host for this virus (Reina et al., 1999). In other reports, PCR followed by restriction enzyme analysis and nucleic acid hybridization were successfully used to detect TYLCV field-infected plants of C. annuum and C. chinense in the Dominican Republic and Cuba (Salati et al., 2002; Quinones et al., 2002) and Jamaica (Roye et al., 1999), respectively. The PCR results showed that most of tested C. frutescens plants were infected with TYLCV. This result supported the previous reports that referred to Capsicum spp. as a host for TYLCV. The first evidence for the susceptibility of C. frutescens, C. baccatum and C. chinense was reported by Polston et al. (2006). Yellowing, mottling and curling symptoms were associated with TYLCV infected pepper plants; however, previous studies stated that the infected plants were symptomless (Morilla et al., 2005; Polston et al., 2006). The observed symptoms may be produced by whitefly feeding damage especially when high numbers of whiteflies were observed on the plants in the field. Because whiteflies are able to transmit this virus from pepper to tomato and vice versa as described by Polston et al. (2006), it may be expected that the virus is transmitted from cultivated tomato of neighboring fields to pepper plants. P. vulgaris is known as a favorable host for many viruses, especially the geminiviruses. The first report of TYLCV infection in P. vulgaris was in Spain (Navas-Castillo et al., 1999). In this study, the observed symptoms of infected plants showed yellowing and light curling unlike those severe curling symptoms described by Kamaal et al. (2015). Perhaps this is because the infection was at the early stage or because our isolate is different. Based on nucleotide sequence and genome length, TYLCV-Pep is shorter than TYLCV-GB and is the same as TYLCV-Genaveh (Lefeuvre et al., 2010), whereas, TYLCV-GB is same as TYLCV-Jazira (Idris and Brown, 2005). The distinction of the genome length occurred in the IR region but not in the protein coding regions because the length of IR of TYLCV-Pep was 290 and 305 bp for the TYLCV-GB. Phylogenetic analysis (Kim et al., 2011) is useful in studying epidemiology and evolutional history of viruses. The phylogenetic tree in Fig. 4 showed that TYLCV-Pep and TYLCV-GB were separated into two clusters. The TYLCV-GB was clustered with TYLCV isolates from Saudi Arabia, Sudan and Yemen while TYLCV-Pep was clustered with isolates from Iran. This result suggests that TYLCV-GB may have originated from Sudan or Yemen but TYLCV-Pep may have originated from Iran. The identity between TYLCV-GB and TYLCV-Pep was 81.2% indicating that they are two deferent strains. Two ORFs were observed on the viral strand of the genome of both viruses. The V1 ORF encodes the coat protein 256 aa in the case of TYLCV-GB and 257 aa in the case of TYLCV-Pep. The coat protein of the virus plays an important role in systemic infection, insect transmission and particle formation (Noris et al., 1998; Kim et al., 2011). The ORF2 V2 encodes a 116 aa in case of TYLCV-GB and 115 aa in case of TYLCV-Pep. V2 protein performs as a suppressor of RNA silencing (Zrachya et al., 2007) and is important for plant infection (Wartig et al., 1997). On the complementary strand of the genome, there are four ORFs in case of TYLCV-Pep and five ORFs in case of TYLCV-GB. These ORFs encoding to C1, C2, C3, C4 and C5 proteins. The C1 ORF encodes the REP (replicase-associated protein) which contains a NTP-binding motif (Laufs et al., 1995) and plays a critical role in viral replication protein. It is 358 aa in both TYLCV-GB and TYLCV-Pep. The C2 protein is a transactivator protein (TrAP) for the expression of virion sense gene and a host range determination factor (Sunter et al., 2001; Wartig et al., 1997). The C2 protein has 134 aa in all TYLCV strains except TYLCV-Pep has 131 aa. The C3 protein has 133 aa; it is known as a replication enhancer (REn) (Settlage et al., 2005), conserved in most TYLCV strains worldwide. The C4 protein is involved in inducing the disease symptoms (Rigden et al., 1994), it has 95 aa in case of TYLCV-GB and 99 aa in case of TYLCV-Pep. The C5 protein (128 aa) is observed in TYLCV-GB and other some TYLCV strains and isolates of Saudi Arabia, Sudan and Yemen (Idris et al., 2012). This potential protein identified on the complimentary strand of TYLCV-GB genome starts from nucleotide 976 to nucleotide 589 and ends with a TGA termination codon. The function of this potential gene is not recognized yet even though it is recorded in some other begomoviruses. The identity of IR sequence between TYLCV-GB and TYLCV-Pep is low (74.6%), TYLCV-GB has 15 nucleotide more than TYLCV-Pep and this may explain the variation of the genome length. The importance of the IR region due to the mutation frequency in this region is higher than the other coding regions. The IR is described as a hot spot for nucleotide disincorporation during the DNA viral replication (Font et al., 2007) and the high level of Begomovirus population diversity might reproduce replication errors (Bull et al., 2006; Zhang et al., 2008). Many of the monopartite begomoviruses have been shown to be associated with two circular ssDNA satellite molecules, betasatellite (DNA-β) and alfasatellite (DNA-1) (Briddon and Stanley, 2006; Fiallo-Olive et al., 2013). The DNA-β depends on their helper virus for replication and encapsidation. Itencodes a gene (βC1) involved in systemic disease development and plays an important role in pathogenicity of their helper virus (Briddon et al., 2003; Fauquet et al., 2008). The DNA-β satellite was found to be associated with TYLCV-GB and shared 90% similarity with the closest relatives that are DNA-β molecules from Yemen (JF919717) (Idris et al., 2012). The amplified DNA-β has one gene (βC1) which encodes a 129 aa protein on the complementary strand. The presence of alfasatellite (DNA-1) did not examine in this research. The first report of DNA-β satellite with TYLCV species was associated with TYLCV from Oman (Khan et al., 2008).
The distinction of genome length, low similarity of genome and IR sequences and the presence of C5 and DNA-β associated with TYLCV-GB but not in TYLCV-Pep-suggest that TYLCV-GB and TYLCV-Pep are two different strains in Saudi Arabia with different invasion routes.
The author gratefully acknowledges the financial support National Plan for Science, Technology and Innovation (MAARIFAH), King Abdullaziz City for Science and Technology, (Grant No. 10-Bio1096-06) and Pests and Plant Disease Unit (PPDU), College of Agricultural and Food Science, King Faisal University provided the support of this study. The author wish to thank many collaborators who have assisted in this study Dr. Khalid Alhudaib, Dr. Hanokh Czosnek and Dr. Jameel Al-Khayri.