Research Article
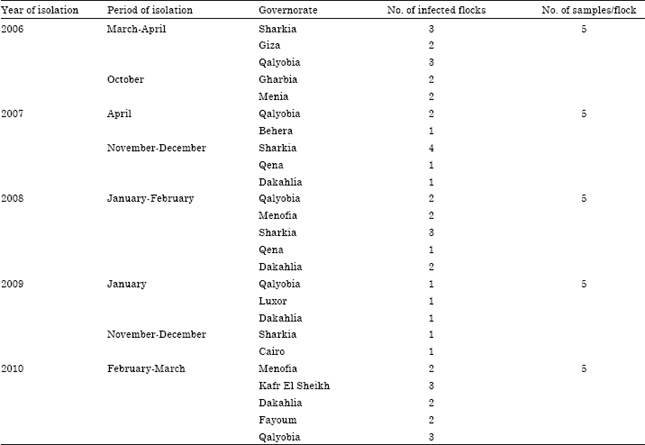
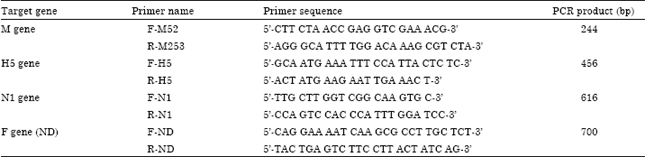
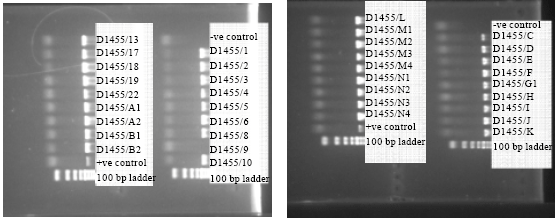
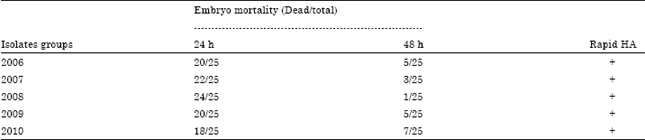
Serological and Molecular Identification of Some Isolated Avian Influenza Viruses During Outbreaks in Egypt
The Central Laboratory for Evaluation of Veterinary Biologics (CLEVB), El Abassia, Cairo, Egypt
Elham A. El-Ibiary
The Central Laboratory for Evaluation of Veterinary Biologics (CLEVB), El Abassia, Cairo, Egypt
A.S. Sadik
Department of Agriculture Microbiology (Virology Laboratory), Faculty of Agriculture, Ain Shams University, Cairo, Egypt
Mamdouh H. Abdel-Ghaffar
Department of Agriculture Microbiology (Virology Laboratory), Faculty of Agriculture, Ain Shams University, Cairo, Egypt
Badwy A. Othman
Department of Agriculture Microbiology (Virology Laboratory), Faculty of Agriculture, Ain Shams University, Cairo, Egypt