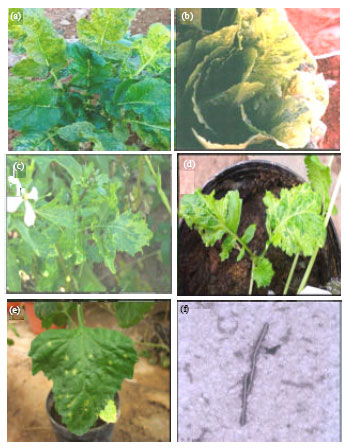
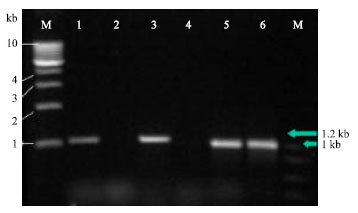
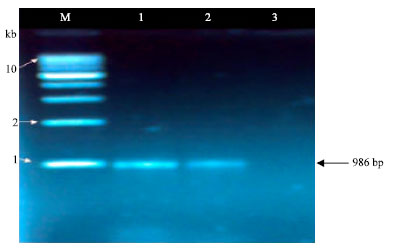
Research Article
Etiology of a Mosaic Disease of Radish and Lettuce and Sequencing of the Coat Protein Gene of the Causal Agent in Saudi Arabia
Department of Plant Protection, College of Food and Agricultural Sciences, King Saud University, P.O. Box 2460, Riyadh 11451, Kingdom of Saudi Arabia
I.M. AL-Shahwan
Department of Plant Protection, College of Food and Agricultural Sciences, King Saud University, P.O. Box 2460, Riyadh 11451, Kingdom of Saudi Arabia
M.A. Amer
Department of Plant Protection, College of Food and Agricultural Sciences, King Saud University, P.O. Box 2460, Riyadh 11451, Kingdom of Saudi Arabia
O.A. Abdalla
Department of Plant Protection, College of Food and Agricultural Sciences, King Saud University, P.O. Box 2460, Riyadh 11451, Kingdom of Saudi Arabia