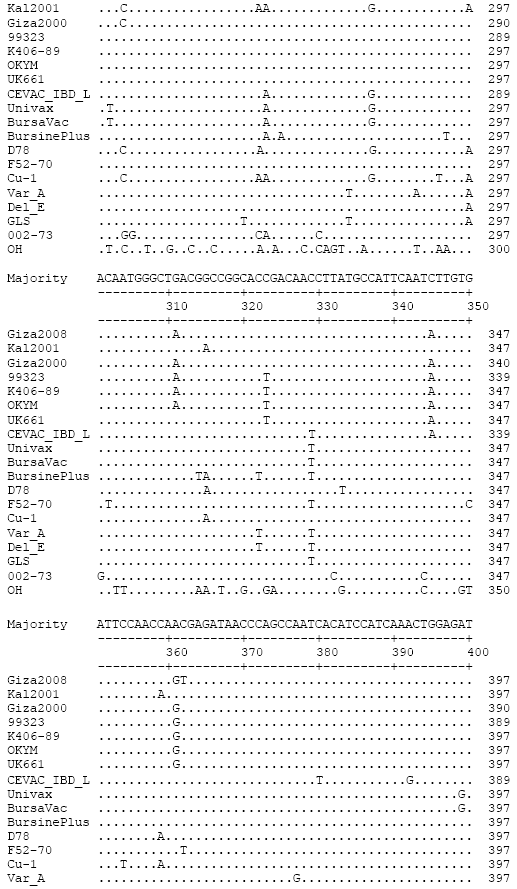
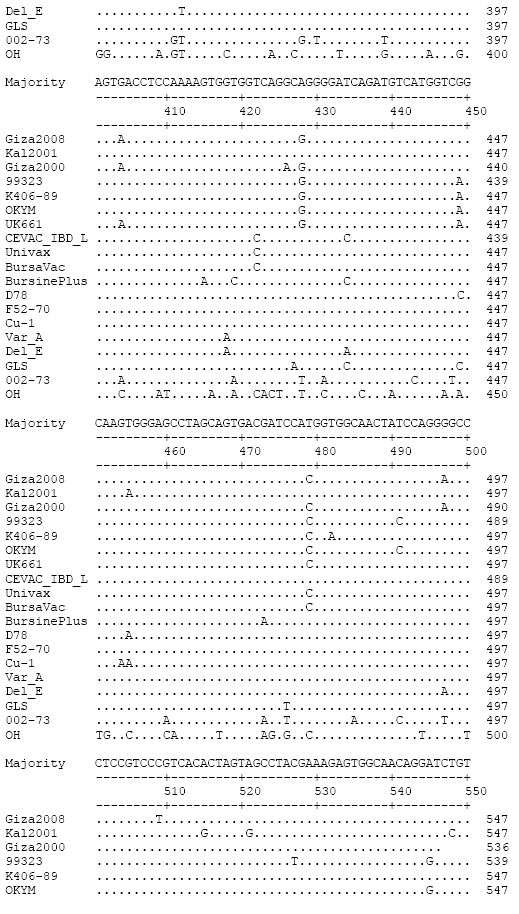
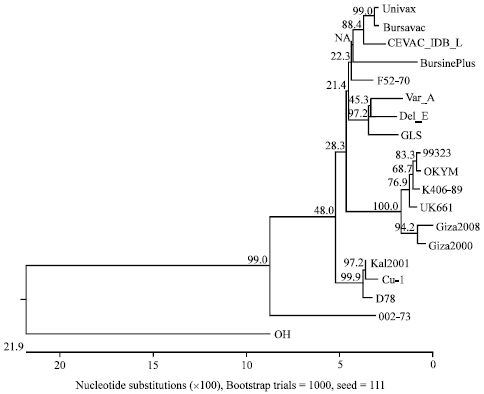
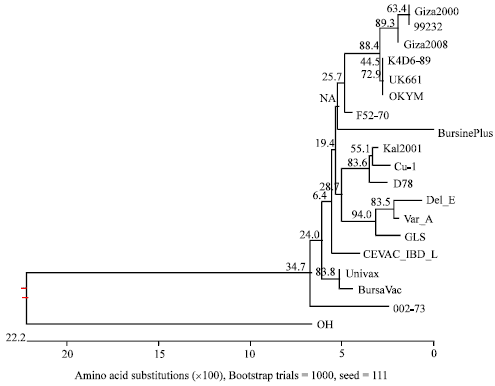
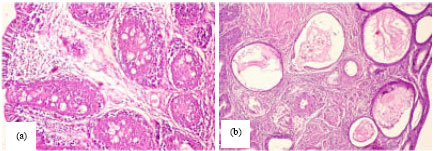
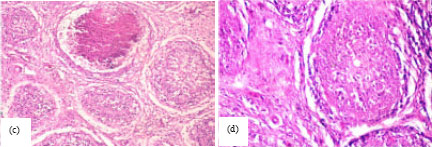
Research Article
Re-Emergence of Very Virulent IBDV in Egypt
Cairo Poultry Company, Giza, Egypt
Ausama A. Yousif
Department of Virology, Faculty of Veterinary Medicine, Cairo University, Egypt
Iman B. Shaheed
Department of Pathology, Faculty of Veterinary Medicine, Cairo University, Egypt
Walaa A. Mohammed
Department of Viral Vaccines of Poultry, Veterinary Serum and Vaccine Research Institute, Abbasia, Egypt
Attia M. Samy
Department of Virology, Faculty of Veterinary Medicine, Cairo University, Egypt
Ismail M. Reda
Department of Virology, Faculty of Veterinary Medicine, Cairo University, Egypt