Research Article
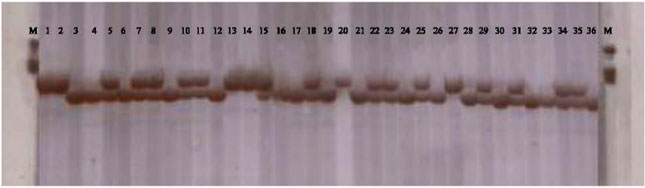
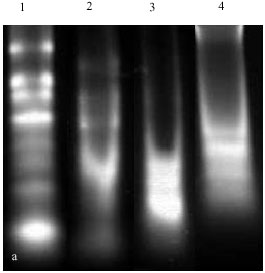
Compared Two Methods for Isolating RNA from Freezing and Nonfreezing Bread Wheat (Triticum aestivum L. ) Plant Tissues
Science and Research Campus, Islamic Azad University of Tehran, Tehran, Iran
R.T. Afshari
Agriculture of College, University of Tehran, Karaj, Iran
H. Alizade
Agriculture of College, University of Tehran, Karaj, Iran
J.T. Afshari
Imonology Research Center, Bu-Ali Institute, Mashhad University of Medical Sciences, Mashhad, Iran
G.R. Javadi
Science and Research Campus, Islamic Azad University of Tehran, Tehran, Iran