Research Article
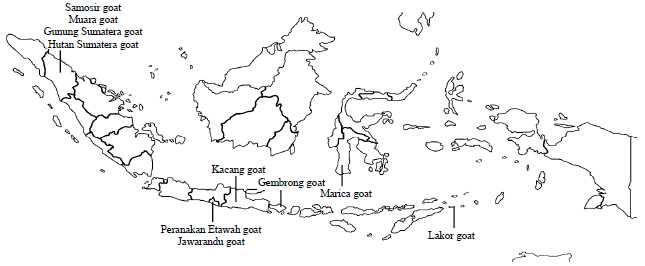
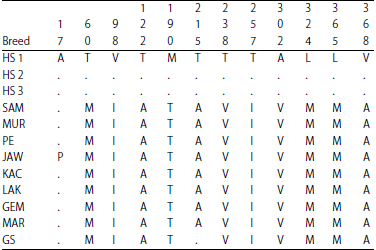
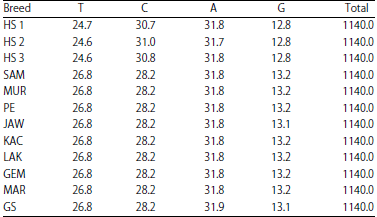
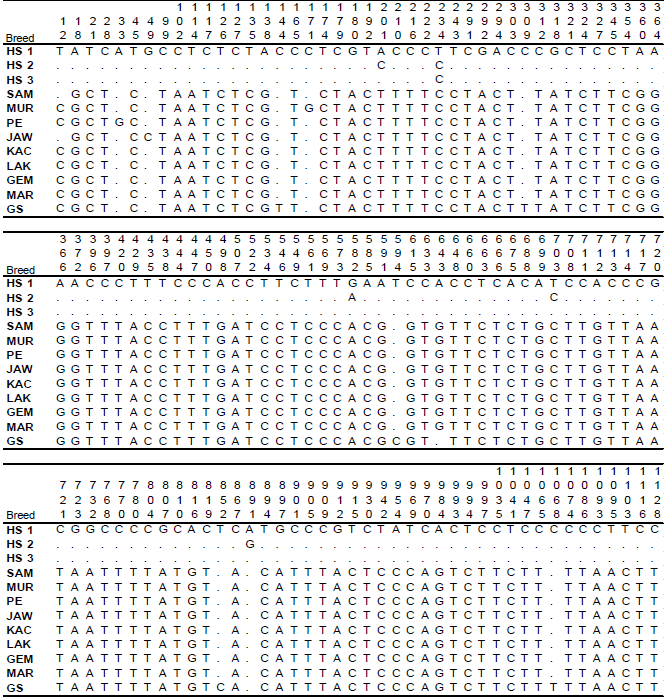
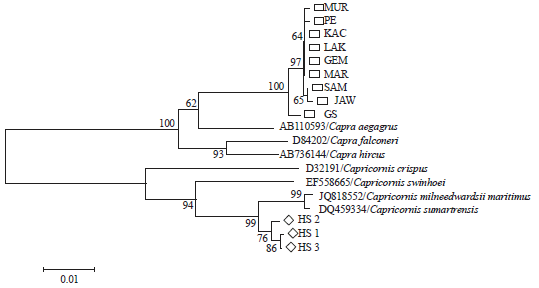
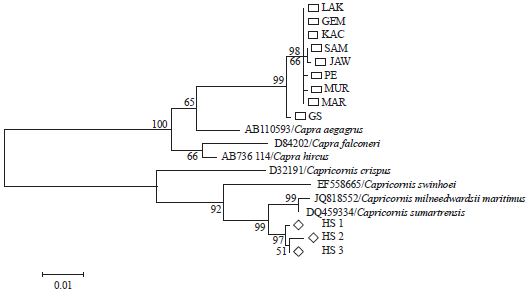
Molecular Phylogenetic of Hutan Sumatera Goat (Sumatran Serow) and Domestic Goat (Capra hircus) in Indonesia Based on Analysis Mitochondrial Cytochrome b Gene
Department of Biochemistry, Faculty of Veterinary Medicine, Gadjah Mada University, 55281 Yogyakarta, Indonesia
Wayan Tunas Artama
Department of Biochemistry, Faculty of Veterinary Medicine, Gadjah Mada University, 55281 Yogyakarta, Indonesia
Rini Widayanti
Department of Biochemistry, Faculty of Veterinary Medicine, Gadjah Mada University, 55281 Yogyakarta, Indonesia
LiveDNA: 62.16531
I. Gede Suparta
Department of Animal Production, Faculty of Animal Science, Gadjah Mada University, 55281 Yogyakarta, Indonesia