Research Article
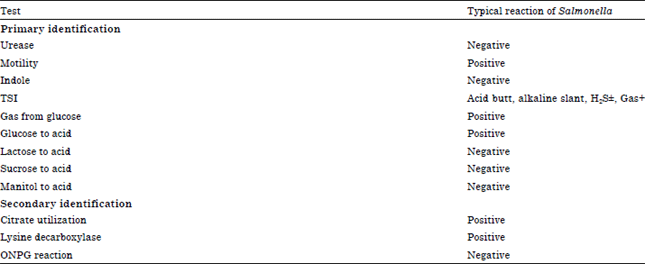
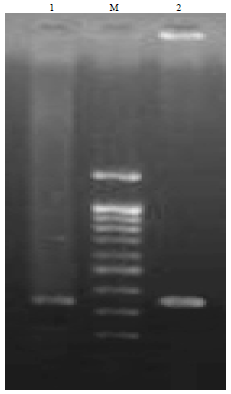
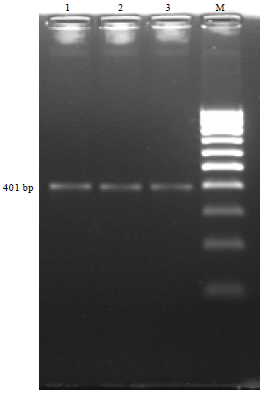
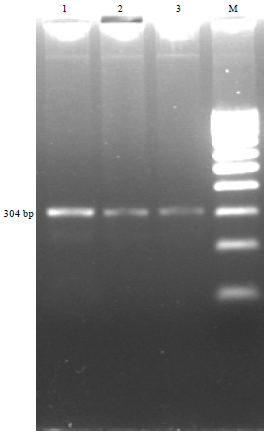
Comparison of PCR and Conventional Cultural Method for Detection of Salmonella from Poultry Blood and Faeces
College of Veterinary and Animal Sciences, Sardar Vallabhbhai Patel University of Agriculture and Technology, Meerut Uttar Pradesh, India
R.K. Agarwal
National Salmonella Center (Veterinary), Division of Bacteriology and Mycology,Izatnagar Uttar Pradesh, India
B. Sailo
Division of Veterinary Public Health,Indian Veterinary Research Institute, Izatnagar Uttar Pradesh, India
M.A. Wani
Division of Veterinary Public Health,Indian Veterinary Research Institute, Izatnagar Uttar Pradesh, India
Ashok Kumar
Division of Veterinary Public Health,Indian Veterinary Research Institute, Izatnagar Uttar Pradesh, India
K. Dhama
Avian Diseases Section, Division of Pathology, Indian Veterinary Research Institute, Izatnagar Uttar Pradesh, India
M.K. Singh
College of Veterinary and Animal Sciences, Sardar Vallabhbhai Patel University of Agriculture and Technology, Meerut Uttar Pradesh, India