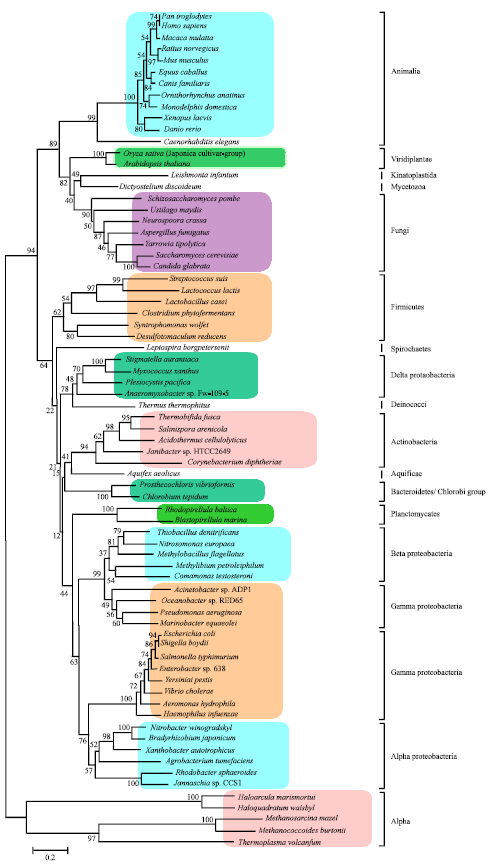
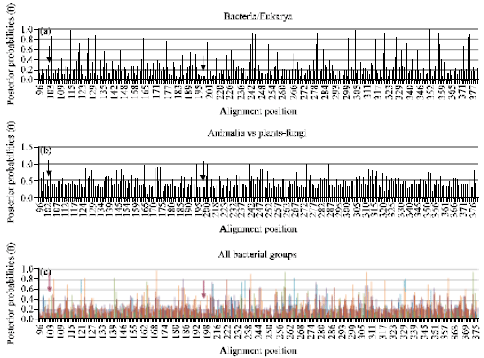
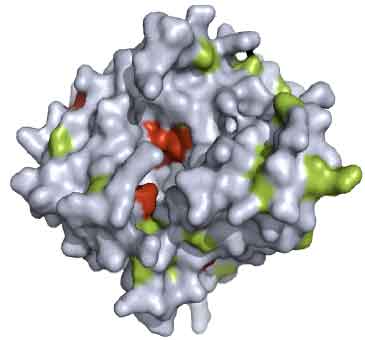
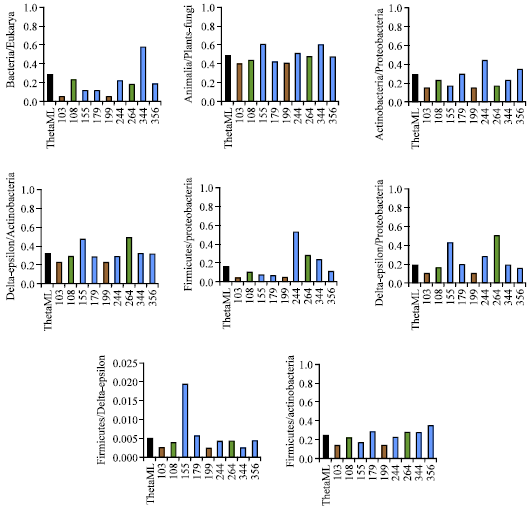
Research Article
Study on the Conserved and the Polymorphic Sites of MTHFR Using Bioinformatics Approaches
Department of Bioinformatics, Research Wing, Indo American Cancer Hospital and Research Center, Hyderabad, India
Kaiser Jamil
Department of Bioinformatics, Research Wing, Indo American Cancer Hospital and Research Center, Hyderabad, India