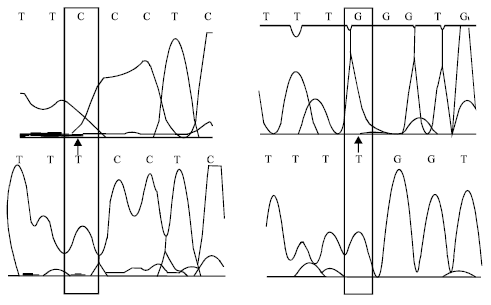
Research Article
Single Nucleotide Polymorphism Associated with Late-onset Alzheimer’s Disease
Department of Zoology, College of Sciences, P. O. Box 2455 King Saud University, Riyadh 11451 Saudi Arabia
Misbahul Arfin
Department of Zoology, College of Sciences, P. O. Box 2455 King Saud University, Riyadh 11451 Saudi Arabia
Bassam A. Alahmadi
Department of Zoology, College of Sciences, P. O. Box 2455 King Saud University, Riyadh 11451 Saudi Arabia
Mohamed A. Al-Jumah
King Fahad National Guard Hospital, Riyadh, Saudi Arabia