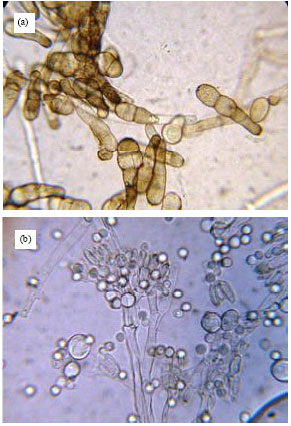
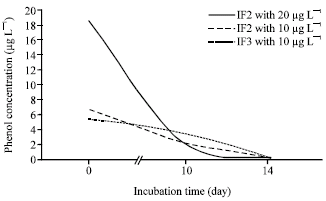
Research Article
Isolation of Two Fungal Strains Capable of Phenol Biodegradation
Department of Biological Sciences, Faculty of Sciences, Al al-Bayt University, Mafraq, Jordan
Sohail Alsohaili
Department of Biological Sciences, Faculty of Sciences, Al al-Bayt University, Mafraq, Jordan