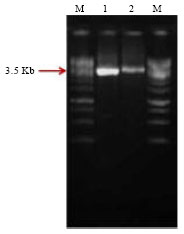
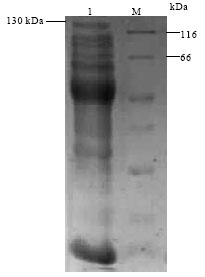
Research Article
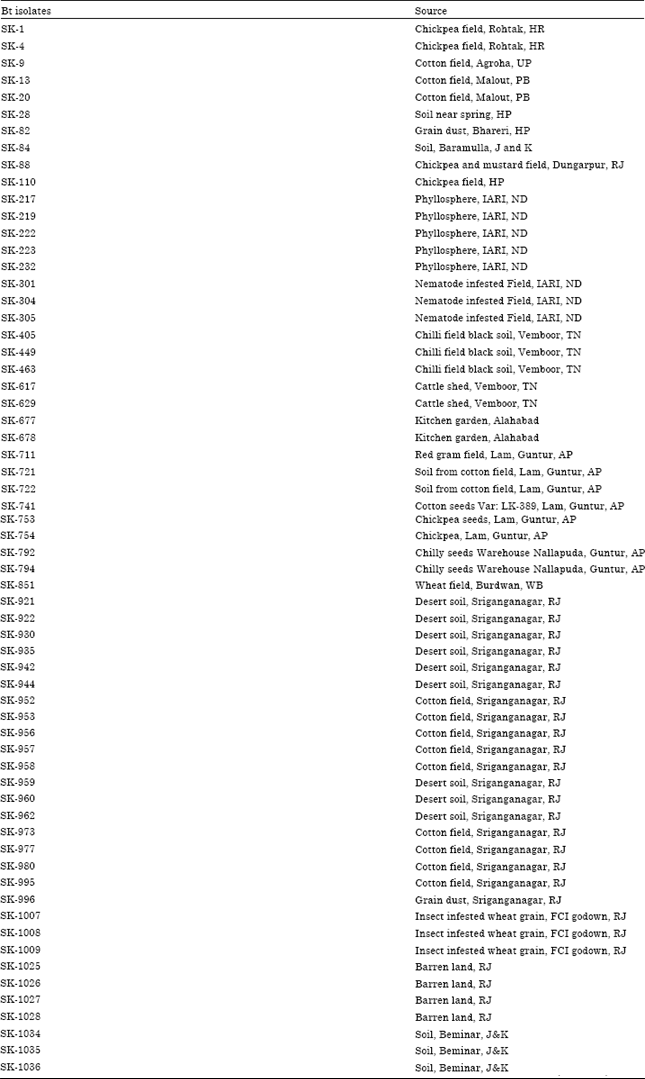
Screening of Bacillus thuringiensis Isolates Recovered from Diverse Habitats in India for the Presence of cry1A-type Genes and Cloning of a cry1Ac33 Gene Toxic to Helicoverpa armigera (American Bollworm)
National Research Centre on Plant Biotechnology, Indian Agricultural Research Institute Campus, New Delhi 110012, India
Gunda Gouthami Krishna Kumari
National Research Centre on Plant Biotechnology, Indian Agricultural Research Institute Campus, New Delhi 110012, India
Alpana Govind
Division of Entomology, Indian Agricultural Research Institute, 110012, New Delhi
T. Gujar
Division of Entomology, Indian Agricultural Research Institute, 110012, New Delhi
Sarvjeet Kaur
National Research Centre on Plant Biotechnology, Indian Agricultural Research Institute Campus, New Delhi 110012, India