Research Article
Genetic Diversity of Lori Goat Population Based on Microsatellite Marker
Department of Genetic, Faculty of Biology, Baku State University, Baku, Azerbaijan
LiveDNA: 98.31936
During the last century, the selection for production traits of the main livestock species has led to a reduction is number of local populations with consequent loss of genetic variability (Sechi et al., 2005). Goats comprise one of the most important domestic livestock species in Iran and play an important role in the livelihood of a larg proportion of small and marginal farmers and landless labourers. Since, the goat provides a good source of meat, milk, fiber and skin, it is popularly known as the poor man's cow. The Lori goats were originally kept in the province of Lorestan. The Lori goats are medium-sized and mostly are black. Natural service is method of Breeding for this goat. The male and female have horns (Fig. 1).
If genetic diversity is very low, none of the individuals in a population may have the characteristics needed to cope with the new environmental conditions or challenges. Such a population could be suddenly wiped out. Low amounts of genetic diversity increase the vulnerability of populations to catastrophic events such as disease outbreaks. Low genetic diversity may also indicate high levels of inbreeding with its associated problems of expression of deleterious alleles or loss of over-dominance. The use of microsatellite regions, segments of the nuclear genom composed of tandem repeats of short sequence motifs, is well established and accepted as a method for the study of genetic information content of animal population (Goldestein and Schlotterer, 2000). A large number of highly polymorphic microsatellites have been characterized and mapped in domestic animals, including sheep, cattle and other ruminants (De-Gortari et al., 1997; Hayes et al., 1996; Jenkins et al., 1997), facilitating the use of these markers for detailed investigation of the genetic structure of a population.
 | |
| Fig. 1: | A Lori goat |
In the case of genetic variability, the analysis of allelic distribution at highly polymorphic microsatellite loci can be used to monitor the genetic structure of populations and to detect changes in the frequency of alleles due to breeding. That also is proven, the microsatellite approach can be applied to the creation of mating schemes aimed to increase genetic variability, reduce consanguinity and enhance fitness within flocks (Tomasco et al., 2002). Microsatellite are highly polymorphic and randomly markers are the simple sequence motif not more than six bases long, that is randomly repeated e.g., (dC-dA)n. Microsatellite being polymorphic, they provide extremely useful markers for comparitive study of genetic variation, parentage contorol, linkage map analysis and could well be the marker of choice for analysis of population structure in domestic species. Microsatellite markers, also known as Simple Sequence Repeats (SSRs) or Short Tandem Repeats (STRs), are regions of DNA that exhibit short repetitive sequence motifs. Because of their high degree of polymorphism, random distribution across the genotypes, microsatellite markers have been proved to be one of the most powerful tools for evaluating genetic diversity and estimating genetic distances among closely populations of ruminant species (Moore et al., 1991; Buchanan et al., 1994; Ellegren et al., 1997). The aim of this study is to investigate the genetic variability of Lori goat population through the analysis of 13 microsatellite markers.
The blood samples were collected from the 46 Animals by puncturing the jugular vein in the vacutainer tubes having EDTA as blood anticoagulant was coll. Then bleeding were transferred them to laboratory (in an ice-cooled box, where they were kept under -20°C in a deep freezer until DNA isolation) and DNA genomic was extracted by salting out method (Miller et al., 1988). We use both spectrophotometry and agarose gel (0.8%) for DNA quality definition. This study was conducted in 2008.
In this study was used 13 microsatellite primer pairs including MAF64, BM4621, BM121, LSCV36, TGLA122, oarJMP23, oarFCB304 ,oarAE133, ILSTS005, ILSTS022, ILSTS029, ILSTS033 and ILSTS34. Most of primers used were independent and belonged to different chromosomes. These loci in prior studies had been amplified on the goat (Maudet et al., 2001; Yang et al., 1999; Hanrahan et al., 1994; Dixit et al., 2008). They showed polymorphism in the goat of world. Thirteen microsatellite markers, their sequences, type of repeat, size rang and their location are shown Table 1.
All PCR reactions were continued the following component: 200 μM dNTPs, 3.5-6 mM MgCl2 , 0.25 μM each of primer, 0.5 unit Taq DNA polymerase, 150 ng DNA. The final volume was 15 μL.
| Table 1: | Microsatellite markers, their sequences, type of repeat, size rang and location |
 | |
| Table 2: | PCR reaction conditions for all loci exceptional TGLA122, oarJMP23 and oarAE133 loci |
 | |
Reactions were run on a thermal cycler (Biometra 96 block T-gradient ,Germany). in this study anealing temperature was modified as following: MAF64 (62.5°C), BM4621 (58°C), LSCV36 (55°C), oar FCB304 (60.5°C) and BM121 (65.5°C). The rest of PCR process is in accordance with the Table 2. For oarJMP23 and TGLA122 primers were used PCR programe (Crawford et al., 1995), for oarAE133 was used PCR programe (Hanrahan et al., 1994) and For ILSTS005, ILSTS022, ILSTS029, ILSTS033 and ILSTS34 primers, The touchdown PCR protocol was used. The alleles and genotypic frequencies directly were identified from the gel (Fig. 2).
Hardy-Weinberg equilibrium (HWE) had been tested based on likelihood ratio for different locus and the number of observed and effective alleles by POPGENE software (Yeh et al., 1999). Polymorphic Information Content (PIC) were estimated by HET software (Ott, 1989).
The PCR reactions were successfully done on all thirteen primers. seven allele in the TGLA122 locus observed in the prior studies on wild goats (capra ibex) but ten allele in Lori goats were observed. For the 13 microsatellites loci analyzed, expected heterozygosity estimates were calculated after Nei (1973), as implement in the POPGENE software to determine genetic variation within the population.
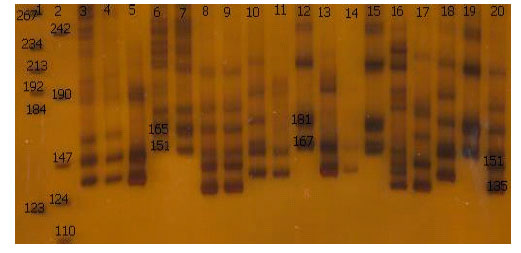 | |
| Fig. 2: | Polyacrylamide nondenaturing gels (8%) showing alleles concerning oarJMP23 marker. DNA size markers are on wells 1, 2 . The alleles and sizes showed in bp |
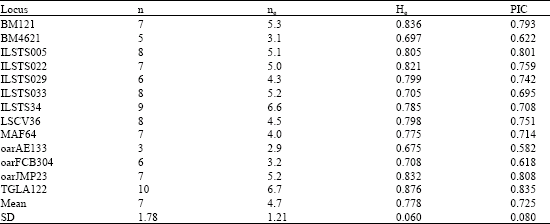
| Table 3: | n, ne, He and PIC values at locus in Lori Goat population |
 | |
Heterozygosity is defined as the probability that a given individual randomly selected from a population will be heterozygous at a given locus. The observed and effective number of alleles was also calculated using POPGENE software (Kimura and Crow, 1964; Yeh et al., 1999). The tests for deviation from Hardy-Weinberg equilibrium were also derived using the exact test of POPGENE.
Number of allele (n), number of allele effective (ne), expected the unbiased average of heterozygosity (He), and Polymorphic Information Content values (PIC) at locus showed in Table 3.
Yang et al. (1999) He value of oarFCB304 locus estimated 0.854 on Chinese goats but it was 0.708 in Lori goat.
Each 13 loci analysis was 100% polymorphic. Highest number of allele objective was 10 allele for TGLA122 locus and lowest number of allele objective was 3 allele for oarAE133 locus. Highest and lowest number of allele effective was 6.7 and 2.9 for TGLA122 and oarAE133 loci with, respectively.
All average the number of allele objective and effective was 7 and 4.70, respectively. Highest and lowest PIC value was 0.835 and 0.582 for TGLA122 and oarAE133, respectively. The average of PIC value for this population was 0.725; it was between 0.746-0.8 in Chinese goats (Yang et al., 1999).
The Lori goats had substantial genetic variation based on their gene diversity and average number of alleles per locus. The average genetic variation (0.778) in Lori goats more than Indian indigenous goats breeds: Barbari, Jamnapari and Sirohi (Ganai and Yadav, 2001). It also demonstrated that microsatellite genotyping is a useful tool for evaluating variation among important goat populations.
Test of genotype frequencies for deviation from HWE at each locus showed this goat population in several loci revealed significant departure from HWE. Deviation from HWE at microsatellite loci have, also been reported in various studies (Barker et al., 2001; Laval et al., 2000; Luikart et al., 1999). It is known that a population is considered to be within HWE only when it is able to maintain its relative allele frequencies. Heterozygosis deficiency is one of the parameters underlying departure from HWE. Heterozygosis deficiency may results from one or more of the following reasons:
| • | The presence of a null allele which is the allele that fails to multiply during PCR using a given microsatellite primer due to a mutation at the primer site (Callen et al., 1993; Pemberton et al., 1995) |
| • | Small sample size, where rare genotypes are likely to be included in the samples; |
| • | The Wahlund effect, i.e., presence of fewer heterozygotes in population than predicted on account of population subdivision |
| • | The decrease in heterozygosity due to increased consanguinity (inbreeding) (Kumar et al., 2006) |
The result of this study suggests that there is substantial genetic variation and polymorphism across the studied loci in Lori goats. The study suggests scope for its further genetic improvement and to undertake appropriate breeding strategies to avoid inbreeding in the population. The information obtained in this study will aid their rational development, utilization and conservation.
This research presents an initial step in investigation of variability at the DNA level within elite breeding flock of Lori goat. The significance of this report is that it offers interesting perspectives for the incorporation of molecular genetic techniques to animal breeding in Iran. In addition the results of this study could provide basic molecular data for the research on germplasm characteristics of Lori goat.
Genetic markers are not only useful for measuring genetic distance between populations but they may also be used in measuring the similarity of individual genotypes with populations. Genetic similarity is a useful method of classifying individuals and populations based on marker genotype information. Further investigation is needed to study the exact properties of this new approach in populations of common origin and inbreed lines over generations.
Author sincere thank to Sanam Montazeri Gharedarvishlu for laboratory practices.