Research Article
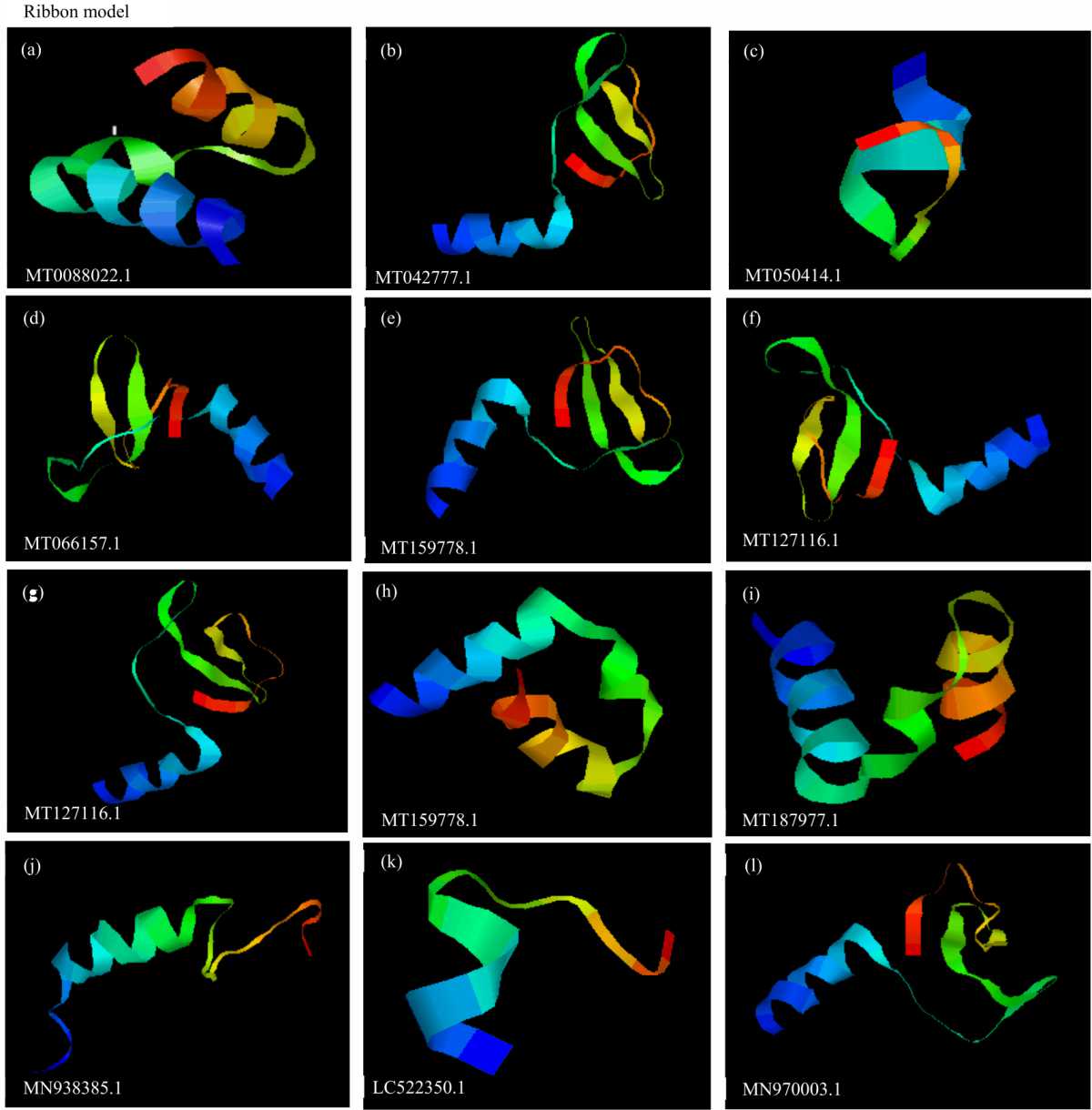
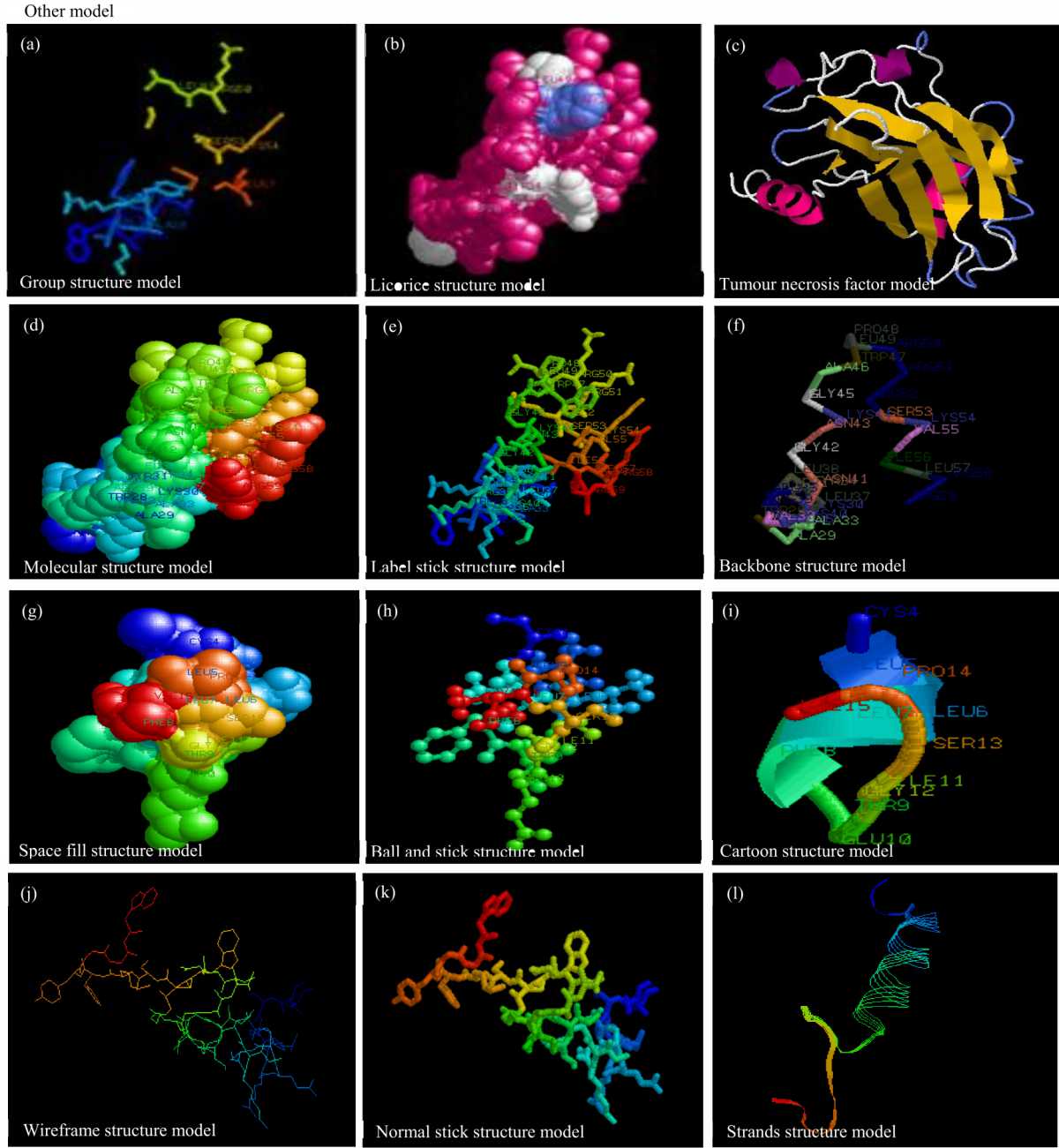
Validation of Isolate Stability Status and in silico Characterization of SARS-COV 2 Partial CdsRdRP Gene Sequences
Department of Genetics and Biotechnology, Faculty of Biological Sciences, University of Calabar, P.M.B 1115, Calabar, Nigeria
LiveDNA: 234.19343
Joseph K. Ebigwai
Department of Plant and Ecological Studies, Faculty of Biological Sciences, University of Calabar, P.M.B 1115, Calabar, Nigeria
LiveDNA: 234.31468
Ndem E. Edu
Department of Genetics and Biotechnology, Faculty of Biological Sciences, University of Calabar, P.M.B 1115, Calabar, Nigeria
Paul B. Ekpo
Department of Genetics and Biotechnology, Faculty of Biological Sciences, University of Calabar, P.M.B 1115, Calabar, Nigeria
Solomon I. Ofem
Department of Microbiology, Faculty of Biological sciences, University of Calabar, P.M.B 1115, Calabar, Nigeria
Imaobong S. Essien
Department of Zoology and Environmental Biology, Faculty of Biological Sciences, University of Calabar, P.M.B 1115, Calabar, Nigeria