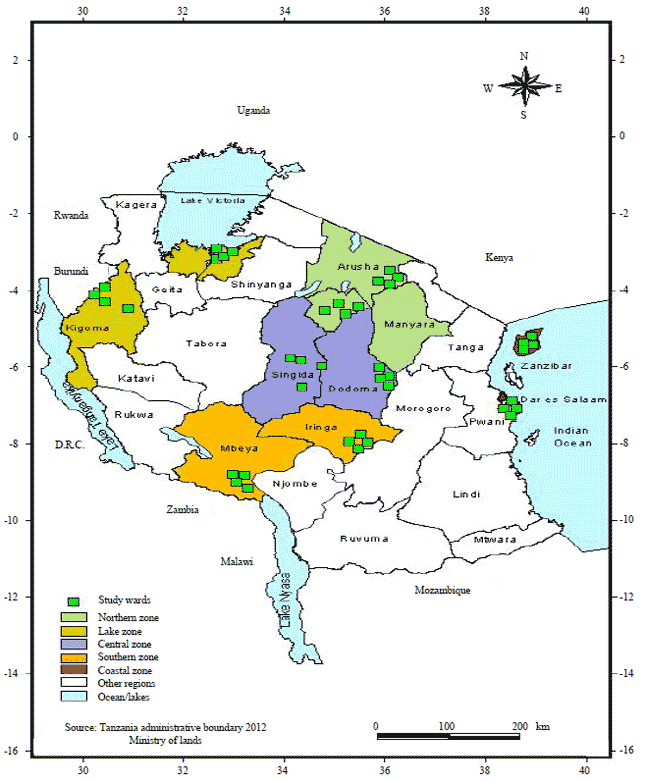
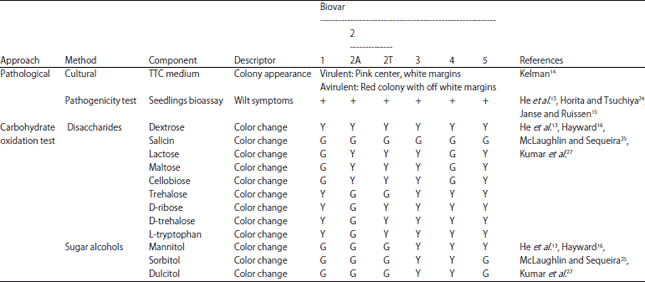
Research Article
Biovar 2 of Ralstonia solanacearum Species Complex Causes Tomato Bacterial Wilt Disease in Tanzania
Department of Sustainable Agriculture and Biodiversity Ecosystem Management, School of Life Sciences and Bioengineering, Nelson Mandela African Institution of Science and Technology, P.O. Box 447, Arusha, Tanzania
LiveDNA: 255.21268
P.A. Ndakidemi
Department of Sustainable Agriculture and Biodiversity Ecosystem Management, School of Life Sciences and Bioengineering, Nelson Mandela African Institution of Science and Technology, P.O. Box 447, Arusha, Tanzania
E.R. Mbega
Department of Sustainable Agriculture and Biodiversity Ecosystem Management, School of Life Sciences and Bioengineering, Nelson Mandela African Institution of Science and Technology, P.O. Box 447, Arusha, Tanzania