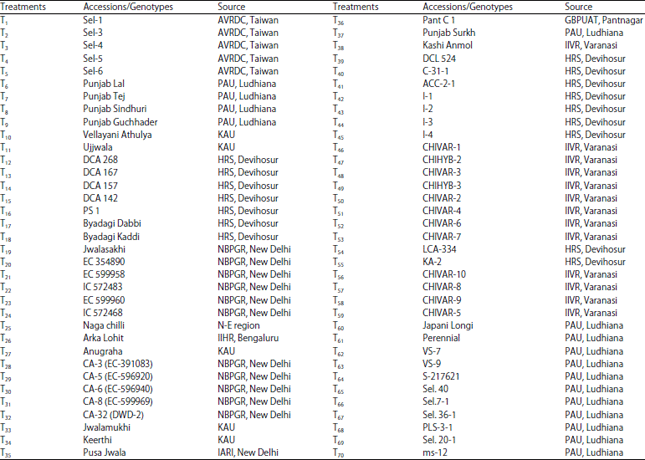
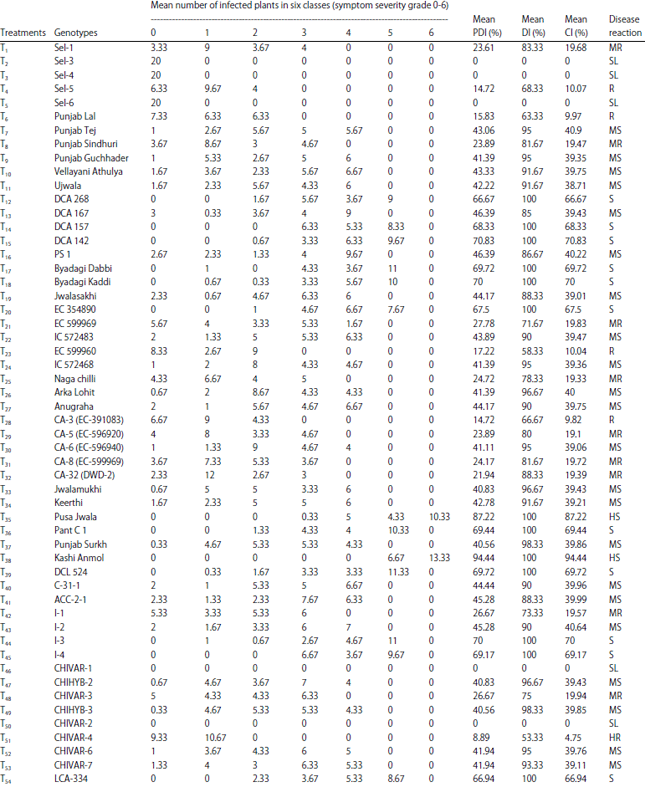
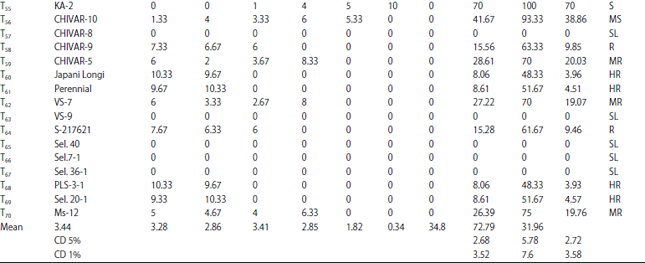
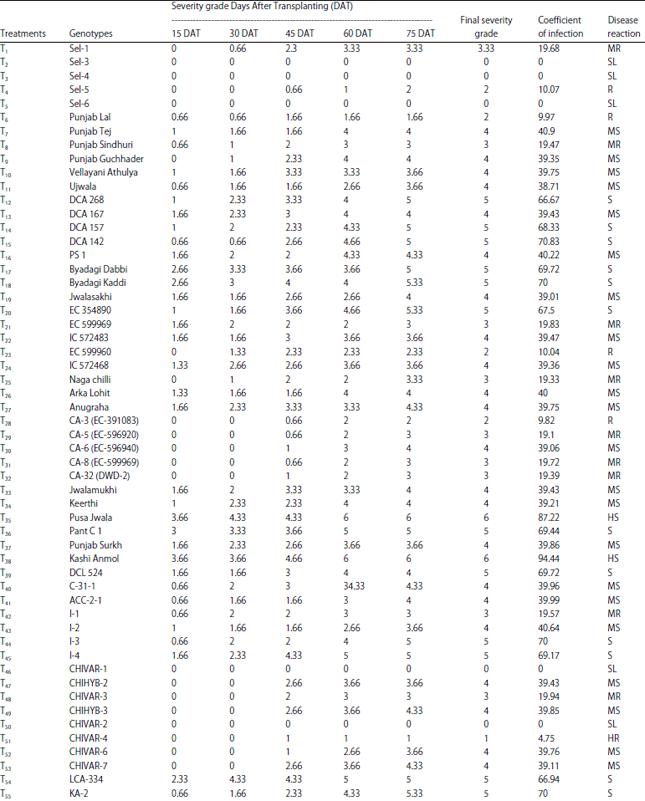
Research Article
Screening Of Popular Indian Chili Pepper (Capsicum annuum L.) Genotypes Against the Chili leaf curl virus Disease
Department of Vegetable Science, College of Agriculture, Kerala Agricultural University, Thrissur, Kerala 680656, India
I. Sreelathakumary
Department of Vegetable Science, College of Agriculture, Kerala Agricultural University, Thrissur, Kerala 680656, India
C. S. Aiswarya
Department of Vegetable Science, College of Agriculture, Kerala Agricultural University, Thrissur, Kerala 680656, India
Prashant Kaushik
Nagano University, 1088 Komaki, Ueda, Nagano 386-0031, Japan