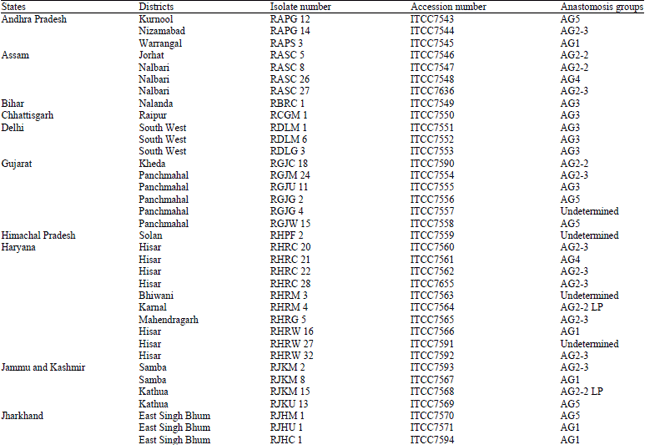
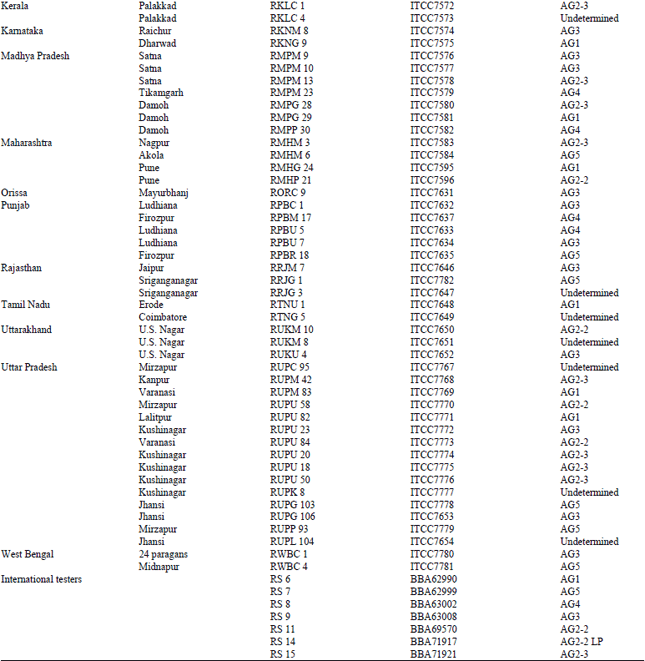
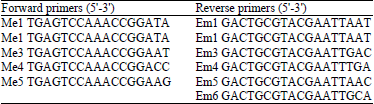
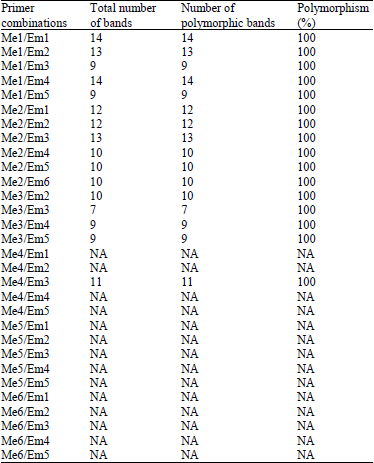
Research Article
Sequence-Related Amplified Polymorphism-PCR Analysis for Genetic Diversity in Rhizoctonia solani Populations Infecting Pulse Crops in Different Agro-Ecological Regions of India
Division of Plant Pathology, ICAR-Indian Agricultural Research Institute, New Delhi, 110012, India
Sunil Chandra Dubey
Division of Plant Quarantine, ICAR-NBPGR, Pusa Campus, New Delhi, 110012, India