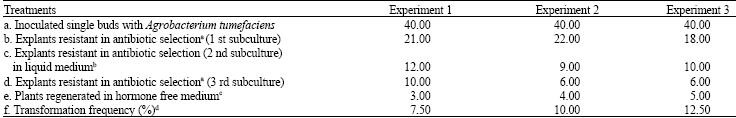
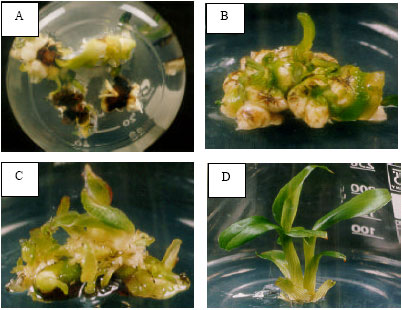
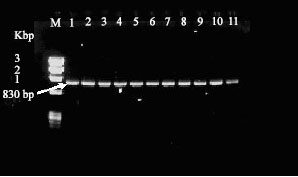
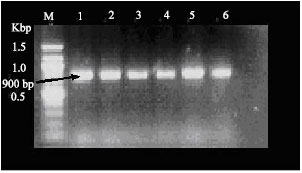
Research Article
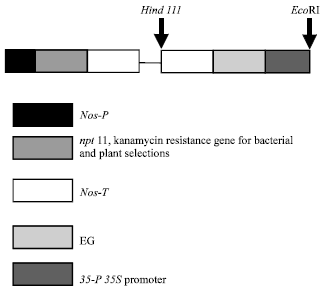
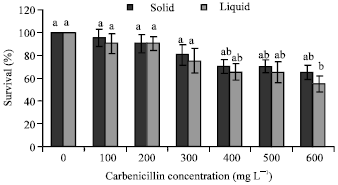
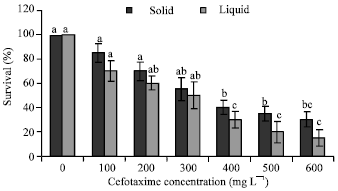
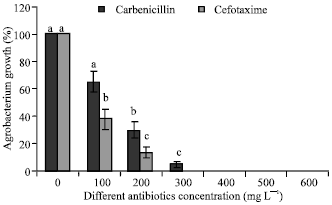
Transgenic Banana Rastali (AAB) with β-1, 3-glucanase Gene for Tolerance to Fusarium Wilt Race 1 Disease via Agrobacterium-mediated Transformation System
Department of Biochemistry, Faculty of Biotechnology and Molecular Sciences, Malaysia
M. Sariah
Department of Plant Protection, Faculty of Agriculture, University Putra Malaysia, 43400, Serdang, Selangor Darul Ehsan, Malaysia
S. Sreeramanan
Department of Biotechnology, Asian Institute of Medicine, Science and Technology (AIMST), Amanjaya, 08000, Sungai Petani, Kedah