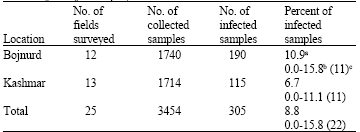
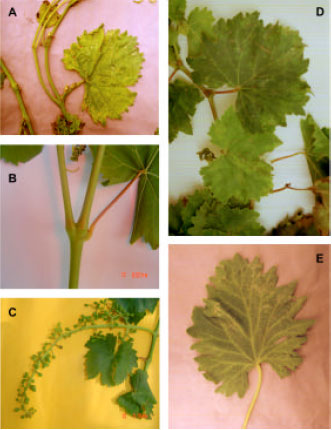
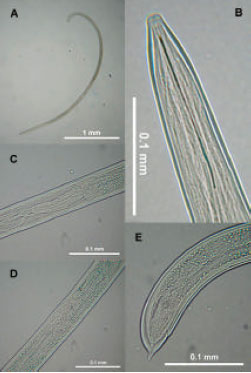
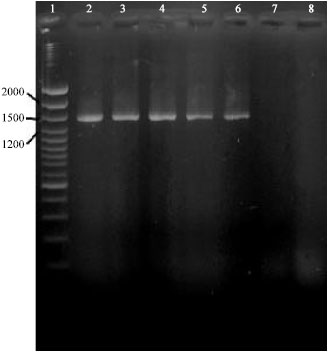
Research Article
Incidence and Distributions of Grapevine fanleaf virus in North-East of Iran
Department of Plant Virology, Plant Pests and Diseases Research Institute, P.O. Box 19395-1454, Tehran, Iran
Shirin Farzadfar
Department of Plant Virology, Plant Pests and Diseases Research Institute, P.O. Box 19395-1454, Tehran, Iran
Ali Reza Golnaraghi
Department of Plant Protection, College of Agriculture and Natural Resources, Science and Research Campus, Islamic Azad University, P.O. Box 14515-775, Tehran, Iran
Ali Ahoonmanesh
Department of Plant Pathology, College of Agriculture, Esfahan University of Technology, Esfahan, Iran