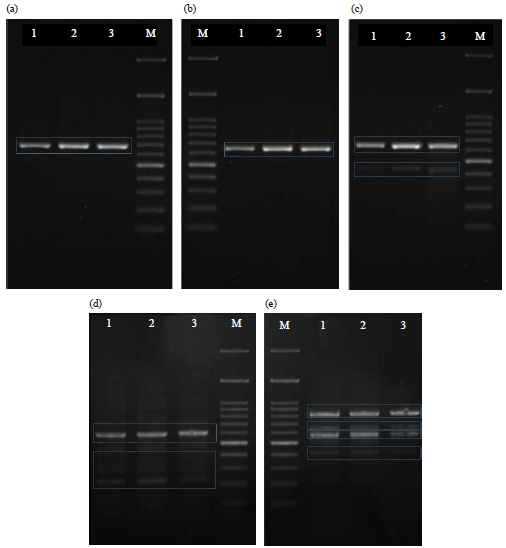
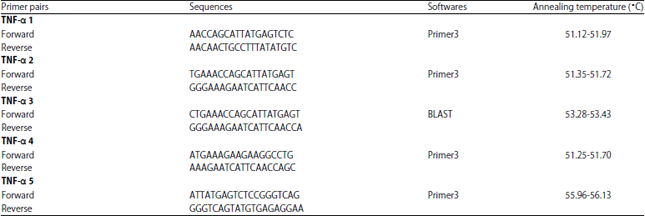
Research Article
Universal Primers for Amplification of TNF-α -308 Promoter Region
Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Sam Ratulangi, Manado, North Sulawesi, Indonesia
Sarah Celia Pertiwi Sumarauw
Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Sam Ratulangi, Manado, North Sulawesi, Indonesia
Sofia Safitri Hessel
Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Sam Ratulangi, Manado, North Sulawesi, Indonesia
Trina Ekawati Tallei
Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Sam Ratulangi, Manado, North Sulawesi, Indonesia
LiveDNA: 62.16548