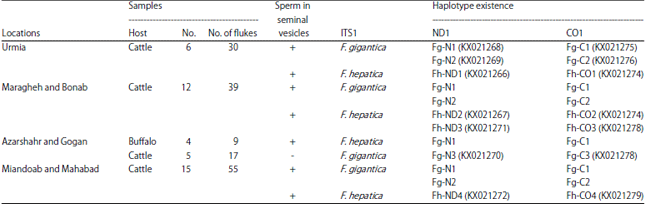
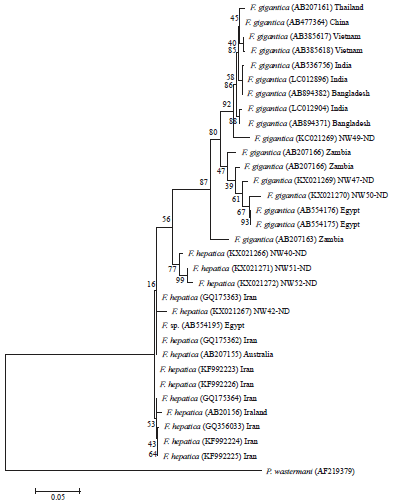
Research Article
Spermatogenic and Phylo-molecular Characterizations of Isolated Fasciola Spp. From Cattle, North West Iran
Department of Parasitology and Mycology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Saber Raeghi
Department of Laboratory Sciences, Maragheh University of Medical Sciences, Maragheh, Iran
Adel Spotin
Department of Parasitology, Faculty of Medical, Tabriz University of Medical Sciences, Tabriz, Iran